About Prof. Dr. J. (Joost) Gribnau
Introduction
Joost Gribnau studied Biochemistry at Leiden University and obtained his PhD in 1999 at the Department of Cell Biology at Erasmus University Rotterdam. He completed his postdoctoral training at the Massachusetts Institute of Technology (Whitehead Institute) and started his own research group in the Department of Cell Biology at Erasmus MC, later moving to the Department of Reproduction and Development. In 2012, he was appointed Professor of Epigenetics, and in 2014 became Head of the Department of Developmental Biology at Erasmus MC.
His research focuses on questions related to sex chromosome and stem cell biology. His group has made important discoveries on the mechanisms driving the female-specific initiation of X-chromosome inactivation and developed the time-machine technology that enables researchers to trace gene and enhancer activity back in time, facilitating whole-genome activity lineage tracing over multiple cell divisions. Together with Dr. Mehrnaz Ghazvini, he established the Erasmus MC iPS Core Facility, where patient-derived stem cells are generated. In 2019, he co-founded Methylomics B.V., a company developing diagnostic tests for cancer detection based on cancer-associated DNA methylation changes. Joost Gribnau received several prestigious grants, including VIDI, VICI, and ERC awards, as well as the Huygens-Descartes Prize. In 2015, he was elected a member of the European Molecular Biology Organization (EMBO).
Fields of expertise
(1) X chromosome inactivation
Group members: Samuel Luchsinger Morcelle, Eveline Rentmeester, Evelyne Wassenaar, Jeffrey Boeren
Many animal species employ sex chromosomes to determine sex and start gender specific gene expression programs. In mammals’ female cells have two X chromosomes, whereas male cells carry an X and Y chromosome. The Y is a small chromosome with not more than 70 coding genes, in contrast to the X chromosome harbouring more than 1000 genes. As a consequence, expression of X linked genes will potentially be two-fold higher in female compared to male cells. Therefore, intricate mechanisms are established to equalize the dosage of X-linked genes between male and female cells. In mammals this involves upregulation of dosage sensitive X-linked genes, and inactivation of one X chromosome in every female somatic cell. X chromosome inactivation (XCI) is regulated by the X-linked X inactivation centre (Xic). This Xic covers a region of ~800kb, and harbours a plethora of long non-coding RNA (lncRNA) genes involved in XCI. Located within the Xic, Xist plays a crucial role in XCI. Xist lncRNA accumulates in cis, thereby recruiting silencing complexes that render the X inactive.

Liver section of a female mouse with X-linked GFP (green) and tdTomato (red) reporters showing patches of cells with exclusive expression of the paternally or maternally inherited X chromosome.
A central question we aim to address in my laboratory is how a cell determines the number of X chromosomes and initiates XCI exclusively in female cells. Our work revealed that initiation of XCI is a stochastic process and indicated the presence of X-linked activators of XCI (Monkhorst 2008). The activity of these XCI-activators is counterbalanced by autosomally encoded XCI-inhibitors, many of which are pluripotency factors involved in repression of Xist, providing a powerful link between loss of pluripotency and XCI initiation. We identified X-encoded RNF12 as an activator of XCI (Jonkers and Barakat 2009). Rnf12 is located just 500 kb telomeric of Xist and encodes an E3 ubiquitin ligase, which catalyzes dose-dependent breakdown of the pluripotency factor REX1 by targeting REX1 for proteasomal degradation (Gontan 2012). When present at an effective concentration, REX1 inhibits Xist transcription, thereby blocking initiation of XCI. Breakdown of REX1 is more prominent in differentiating female cells, which still have two active copies of Rnf12, resulting in female specific initiation of XCI. The REX1-RNF12 axis provides a strong link between female specific initiation of XCI and loss of pluripotency, but our studies and studies of others also indicated the presence of more XCI-activators (Barakat 2015, Gontan 2018). To identify these, we performed a CRISPR/Cas9 screen targeting large X-chromosomal regions, which revealed Hcfc1 as another X-linked XCI activator. HCFC1 is member of the COMPASS complex that is recruited by YY1 to Xist regulatory regions leading to increased levels of histone modification H3K4me3 and transcription activation. Further analysis showed that besides recruitment of HCFC1 and COMPASS, YY1 also plays a central role in recruiting the Integrator complex genome-wide, including at Xist regulatory sites, highlighting how novel insights in mechanisms governing XCI contribute broadly to our understanding of gene regulation in general. Our present research is aimed at identifying novel XCI-activators, elucidate the mechanisms by which they direct female exclusive XCI, and translate these findings to human.
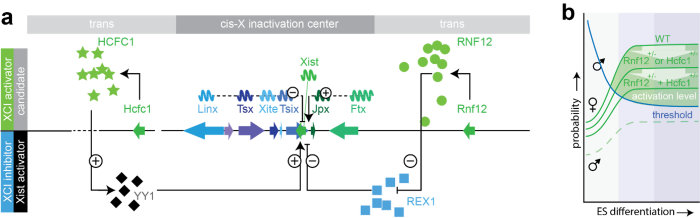
XCI regulatory network. a) Xist regulatory network of known Xist cis-regulatory genes (arrows depict genes: positive regulators of Xist in green, negative in blue), and trans-acting factors (X-linked XCI-activators in green, autosomal encoded Xist-activators and XCI inhibitors in black and blue, respectively). Long non-coding RNA genes are indicated with waves. b) Only in female cells is the concentration of XCI activators sufficient to reach the threshold required for Xist activation. This activation level is reduced in female cells carrying heterozygous deletions of the XCI activators Rnf12 and/or Hcfc1.
Relevant publications:
-Allsop R*, Boeren J*, Tan B*, Merzouk S, van Ijcken W, Mira Bontenbal H, Gontan C, Gribnau J#, Pasque V#. 2025 X-chromosome upregulation operates on a gene-by-gene basis at RNA and protein levels. Nature Communications. In press.
-Robert-Finestra T, Tan B, Mira-Bontenbal H, Timmers E, Gontan C, Merzouk S, Giaimo B, Dossin F, van IJcken W, Martens J, Borggrefe T, Heard E, and Gribnau J. SPEN is required for Xist upregulation during initiation of X chromosome inactivation. Nature Communications. 2021.
-van Bemmel JG, Gard C, Galupa R, Servant N, Picard C, Davies J, Szempruch T, Zhan Y, Baulande S, Gentien S, Nora EP, Giorgetti L, Guttman M, Hughes J, Higgs D, Gribnau J, Heard E. Inverting the TAD boundary at the antisense Xist/Tsix locus leads to Xist and Tsix misregulation at the onset of X-inactivation. Nature Genetics. 2019.
-Gontan C, Mira-Bontenbal H, Magaraki A, Dupont C, Barakat TS, Rentmeester E, Demmers J, Gribnau J. REX1 is the critical target of RNF12 in imprinted X chromosome inactivation in mice. Nat Communications. 2018
-Barakat TS, Loos F, van Staveren S, Myronova E, Ghazvini M, Grootegoed JA, Gribnau J. The trans-activator RNF12 and cis-acting elements effectuate X chromosome inactivation independent of X-pairing. Mol Cell. 2014.
-Gontan C, Achame EM, Demmers J, Barakat TS, Rentmeester E, van IW, Grootegoed JA, Gribnau J. RNF12 initiates X-chromosome inactivation by targeting REX1 for degradation. Nature. 2012.
Jonkers I*, Barakat TS*, Achame EM, Monkhorst K, Kenter A, Rentmeester E, Grosveld F, Grootegoed JA, Gribnau J. RNF12 is an X-Encoded dose-dependent activator of X chromosome inactivation. Cell. 2009.
-Monkhorst K, Jonkers I, Rentmeester E, Grosveld F, Gribnau J. X inactivation counting and choice is a stochastic process: evidence for involvement of an X-linked activator. Cell. 2008.
Group members: Beatrice Tan, Joachim Boers, Ruben Boers, Marieke van Leeuwen
The development of more than 300 cell types present in our body requires the action of complex molecular mechanisms, involving signal transduction pathways that instruct gene transcription by impacting on transcription factor networks and the epigenetic landscape. Epigenetic mechanisms play a crucial role in installing and maintaining cell fate decisions. Our interest and expertise in embryonic stem cells, which we use as a model system to study XCI, has sparked research lines involving addressing research questions related to stem cell biology. For instance, my laboratory provided critical knowledge and infrastructure to setup organoid studies which were performed in close collaboration with clinical research groups, either through ESC or iPSC differentiation to kidney or forebrain organoids (Gomes 2020, Shankar 2022) or by expansion of tissue into kidney or placental organoids (Xu 2022, Schäffers 2022). We recently developed ETX embryo’s through self-assembly of different stem cell lines (ESC, TSC, and XEN, Dupont 2023). These models resemble post-implantation egg cylinder in vivo embryo’s providing a powerful model system to study embryonic development in vitro as well as XCI. To better understand cell state changes in development and cell differentiation we developed the DCM time machine (DCM-TM) technology (Boers 2023). DCM-TM enables the reconstruction of gene and enhancer activity maps of the past. DCM-TM utilizes an inducible DCM-RNA polymerase subunit-b fusion protein to label active genes and enhancers with a bacterial methylation mark. This mark does not interfere with transcription and is stably propagated in S-phase. We applied DCM-TM to study intestinal homeostasis, revealing new insights into cell signalling and chromatin and transcription factor activity dynamics in stem cell differentiation. DCM-TM is a versatile tool that can be applied to follow cell differentiation, embryonic development and tissue regeneration, identifying temporal maps of transcription factor networks and signal transduction pathways that can be utilized to improve stem cell expansion and cell differentiation models.
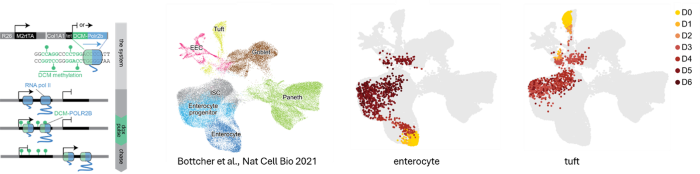
The DCM-TM technology. DCM-TM experimental setup (left), where DCM labelling of gene bodies is induced and maintained over multiple cell divisions. The right panels show annotation of cell types in a scRNA-seq UMAP (Bottcher) and projection of cell state changes observed with DCM-TM with enterocyte and tuft cell viewpoints (in days chase).
Relevant publications:
-Boers R, Boers J, Tan B, van Leeuwen M, Wassenaar E, Gonzalez Sanchez E, Sleddens E, Tenhagen Y, Mulugeta E, Laven J, Creyghton M, Baarends W, van IJcken W, Gribnau J. Retrospective analysis of enhancer activity and transcriptome history. Nature Biotechnology. 2023.
-Dupont C, Schäffers OJM, Tan BF, Merzouk S, Bindels EM, Zwijsen A, Huylebroeck D, Gribnau J. Efficient generation of ETX embryoids that recapitulate the entire window of murine egg cylinder development. Science Adv. 2023.
-Xu Y, Kuppe C, Perales-Patón J, Hayat S, Kranz J, Abdallah AT, Nagai J, Li Z, Peisker F, Saritas T, Halder M, Menzel S, Hoeft K, Kenter A, Kim H, van Roeyen CRC, Lehrke M, Moellmann J, Speer T, Buhl EM, Hoogenboezem R, Boor P, Jansen J, Knopp C, Kurth I, Smeets B, Bindels E, Reinders MEJ, Baan C, Gribnau J, Hoorn EJ, Steffens J, Huber TB, Costa I, Floege J, Schneider RK, Saez-Rodriguez J, Freedman BS, Kramann R. Adult human kidney organoids originate from CD24+ cells and represent an advanced model for adult polycystic kidney disease. Nat Genetics. 2022
-Shankar AS, Du Z, Tejeda Mora H, Boers R, Cao W, van den Bosch TPP, Korevaar SS, Boers J, van IJcken WFJ, Bindels EMJ, Eussen B, de Klein A, Pan Q, Oudijk L, Clahsen-van Groningen MC, Hoorn EJ, Baan CC, Gribnau J, Hoogduijn MJ. Kidney Organoids Are Capable of Forming Tumors, but Not Teratomas. Stem Cells. 2022
-Gomes AR, Fernandes TG, Vaz SH, Silva TP, Bekman EP, Xapelli S, Duarte S, Ghazvini M, Gribnau J, Muotri AR, Trujillo CA, Sebastião AM, Cabral JMS, Diogo MM. Modeling Rett Syndrome With Human Patient-Specific Forebrain Organoids. Front Cell Dev Biol. 2020
-Gunhanlar N, Shpak G, van der Kroeg M, Gouty-Colomer LA, Munshi ST, Lendemeijer B, Ghazvini M, Dupont C, Hoogendijk WJG, Gribnau J, de Vrij FMS, Kushner SA. A simplified protocol for differentiation of electrophysiologically mature neuronal networks from human induced pluripotent stem cells. Mol Psychiatry. 2017.
-Dupont C, Loos F, Kong ASJ, Gribnau J. FGF treatment of host embryos injected with ES cells increases rates of chimaerism. Transgenic Res. 2017.
-Barakat TS, Ghazvini M, de Hoon B, Li T, Eussen B, Douben H, van der Linden R, van der Stap N, Boter M, Laven JS et al. Stable X chromosome reactivation in female human induced pluripotent stem cells. Stem Cell Reports. 2015.
As part of the development of the DCM time machine technology and to be able to trace the DCM methylation labels at very low cost we developed the Methylated DNA sequencing technology (MeD-seq, Boers Genome Res. 2018, Boers Nature biotech. 2023). MeD-seq is based on a methylation dependent restriction enzyme LpnPI which generates 32 base pair fragments around a fully methylated target sequence. Besides DCM methylation MeD-seq detects 50% of all methylated CpG sequences genome wide and can be applied on low amounts of input DNA (<2.5ng or <500 cells), as well as on laser dissected FFPE treated material and cell free DNA (cfDNA). Consequently, MeD-seq is used in many collaborations with clinicians to study disease (>60 collaborations of which >20 published so far). These studies indicate the enormous potential of cfDNA applications to study disease and treatment success remotely. Applications are not restricted to cancer diagnosis and treatment monitoring alone but also include early detection of FTD and monitoring of placental growth and detection of preeclampsia at an early stage (Deger Clin. Epigenetics 2021; Giannini Ann. Clin. Transl. Neurol. 2024; van Vliet Mol. Hum. Reprod. 2025).
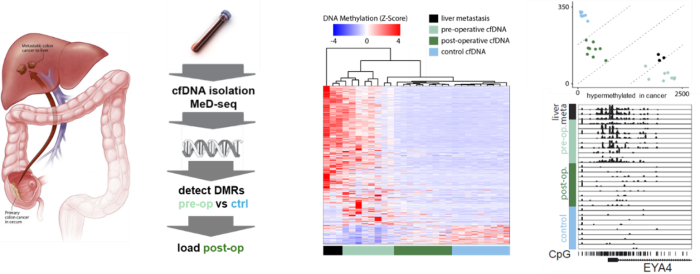
Figure; cfDNA of controls and pre- and post-operative patients with metastasized colon cancer to the liver were subjected to MeD-seq showing loss of the cancer specific DNA methylation profile in post-operative patients (middle panel), that can be quantified (upper right panel). EYA4 is a marker specifically methylated in colon cancer (right bottom panel, collaboration with Saskia Wilting and John Martens, department of Medical Oncology).
Relevant publications:
-van den Berg CB, Nieuwenhuyzen-de Boer GM, Boere IA, Boers RG, Boers JB, van-IJcken WFJ, Jansen MPHM, Kirmizitas TS, Gribnau JH, van Beekhuizen HJ. Genome-wide cell-free DNA methylation profiling in advanced stage ovarian cancer. Are we looking at the tumor or the patient's immune response to the tumor? Cancer Treat Res Commun. 2025
-van Vliet MM, Boers RG, Boers JB, Schäffers OJM, van der Meeren LE, Gribnau J, Schoenmakers S, Steegers-Theunissen RPM. Profiling (placental) DNA methylation in cell-free DNA across gestation: the Rotterdam Periconception Cohort. Mol Hum Reprod. 2025
-Giannini LAA, Boers RG, van der Ende EL, Poos JM, Jiskoot LC, Boers JB, van IJcken WFJ, Dopper EG, Pijnenburg YAL, Seelaar H, Meeter LH, van Rooij JGJ, Scheper W, Gribnau J, van Swieten JC. Distinctive cell-free DNA methylation characterizes presymptomatic genetic frontotemporal dementia. Ann Clin Transl Neurol. 2024
-Boers R, Boers J, Tan B, van Leeuwen M, Wassenaar E, Gonzalez Sanchez E, Sleddens E, Tenhagen Y, Mulugeta E, Laven J, Creyghton M, Baarends W, van IJcken W, Gribnau J. Retrospective analysis of enhancer activity and transcriptome history. Nature Biotechnology. 2023.
-Smit KN, Boers R, Vaarwater J, Boers J, Brands T, Mensink H, Verdijk RM, van IJcken WFJ, Gribnau J, de Klein A, Kilic E. Genome-wide aberrant methylation in primary metastatic UM and their matched metastases. Sci Rep. 2022
-Deger T, Boers RG, de Weerd V, Angus L, van der Put MMJ, Boers JB, Azmani Z, van IJcken WFJ, Grünhagen DJ, van Dessel LF, Lolkema MPJK, Verhoef C, Sleijfer S, Martens JWM, Gribnau J, Wilting SM. High-throughput and affordable genome-wide methylation profiling of circulating cell-free DNA by methylated DNA sequencing (MeD-seq) of LpnPI digested fragments. Clin Epigenetics. 2021
-Slot E, Boers R, Boers J, van IJcken WFJ, Tibboel D, Gribnau J, Rottier R, de Klein A. Genome wide DNA methylation analysis of alveolar capillary dysplasia lung tissue reveals aberrant methylation of genes involved in development including the FOXF1 locus. Clin Epigenetics. 2021
-Boers R, Boers J, de Hoon B, Kockx C, Ozgur Z, Molijn A, van IJcken W, Laven J, Gribnau J.Genome-wide DNA methylation profiling using the methylation-dependent restriction enzyme LpnPI. Genome Res. 2018
