About Dr. G. (Gert) Jansen
Introduction
Gert Jansen received his PhD at the Dept. of Cell Biology, Nijmegen University, where he studied the molecular basis of the human disease Myotonic Dystrophy. He did his postdoctoral training at the Netherlands Cancer Institute in Amsterdam where he studied the PIM kinases in the nematode C. elegans, and developed a new method to inactivate genes in C. elegans. Subsequently, as a PULS post-doctoral fellow, he used the new gene-inactivation method to analyze the functions of all heterotrimeric G proteins (21 Gα, 2 Gβ and 2 Gγ genes) in C. elegans. In 2000 he continued the analysis of G protein signaling as a "young promising scientist" appointed by the Centre for Biomedical Genetics at the Department of Cell Biology and Genetics at the Erasmus MC. He received a KNAW fellowship to study the molecular mechanisms that regulate the behavioral response to salts in the nematode C. elegans.
Research in the Jansen laboratory originally focused on heterotrimeric G protein signaling in the sensory neurons of C. elegans. This has resulted in papers on the roles of different Gα subunits in olfaction (Lans et al., 2004), in the regulation of gene expression in the olfactory neurons (Lans et al., 2006) and the regulation of longevity by sensory G proteins (Lans et al., 2007 & 2009). In addition, in collaboration with Drs. Teng and McCafferty, the Jansen lab developed a C. elegans based system to identify and characterize GPCR-ligand pairs (Teng et al., 2006 & 2008).
Currently the work in the Jansen lab focuses on how cells and organisms sense their environment. Using a new paradigm to analyze behavioral plasticity, where worms learn to avoid NaCl, we identified cells and genes involved and showed that the animals modulate the activity of the sensory neurons, allowing them to switch between attraction to and avoidance of NaCl, depending on their previous experience (Hukema et al., 2006 & 2008; Janssen et al., 2008 & 2009; Beets et al., 2012; Dekkers et al., 2021).
In addition, we use the nematode C. elegans and cultured mammalian cells to identify proteins that regulate cilium length (Burghoorn et al., 2007 & 2010; Broekhuis et al., 2013 & 2014; Van der Vaart et al., 2015; Kunova Bosakova et al., 2018 & 2019) and characterized the functions of several ciliary proteins (Timbers et al., 2016; Loucks et al., 2016; Kenter et al., 2019 & 2020; Van der Burght et al., 2020; Chrystal et al., 2022).
Finally, we use C. elegans to unravel the molecular mechanisms that maintain cell fate Gonzaloz Barrios et al., 2015; Traets et al., 2021). We uncovered a novel mechanism that involves preferential binding of the terminal selector transcription factor CHE-1 to its own promoter over binding to the promoters of its target genes.
Contact information
E-mail: g.jansen@erasmusmc.nl
Telephone office: +31-107043473
Telephone lab: +31-10-7043165
Visiting address
Dept. of Developmental Biology
Erasmus MC
Faculty building
Room Ee-1073
Dr. Molewaterplein 40
3015 GD Rotterdam
The Netherlands
Mail address
Dept. of Developmental Biology
Erasmus MC
PO Box 2040
3000 CA Rotterdam
The Netherlands
Research interests
How do cells and organisms sense their environment?
Accurate detection of cues in the environment is essential for the survival of all organisms. The same holds true for all cells. Based on signals from their environment and internal cues, cells decide to divide, migrate, differentiate, live or die. Each cell or cell type expresses a particular set of receptors and channels, setting their cell fate and making them sensitive to particular cues. Many receptors and channels are located on the plasma membrane of the cell, but some localize to a specialized sensory organelle, the primary cilium.
Our interests can be divided into four subjects:
- Primary cilia: sensory organelles
- Sensory signal transduction, focusing on salt (NaCl) taste
- Maintenance of cell fate
- C. elegans as a model organism
Primary cilia: sensory organelles
Primary cilia are present on nearly all cells of the vertebrate body and extend from the cell’s surface. They harbor specific receptors, channels and other signaling molecules depending on the cell type. Important examples of signaling routes that use cilia are Hh, Wnt, PDGF, IGF, EGF, FGF, TGFβ, Notch, vasopressin, polycystin, somatostatin, serotonin and dopamine signaling.
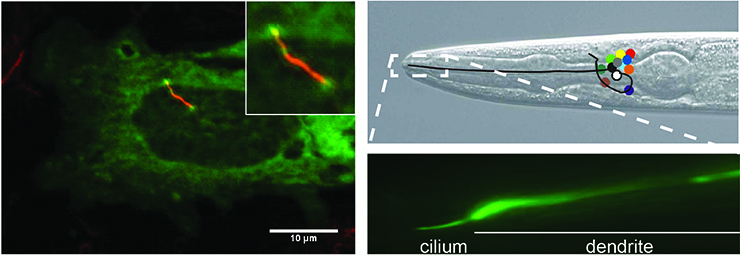
Left panel: Cilium of IMCD3 cell visualized by anti-acetylated tubulin staining (red).
Right panels: Head of C. elegans and cilium of one of the sensory neurons visualized using GFP. Positions of cell bodies of neurons are indicated with colored circles.
Given the large number of signaling routes that use cilia and their presence on almost all vertebrate cells, it is not surprising that cilia dysfunction is the cause of many diseases and can result in different symptoms including infertility, polydactyly, retina degeneration, mental retardation, obesity and kidney cyst formation. Together these diseases are called ciliopathies.
We use the nematode C. elegans and cultured mammalian cells to study the interplay between cilia and signaling. On the one hand, we would like to understand how cilia are build and how their length and morphology are regulated, among others by environmental signals. On the other hand, we study cilium function, how cilia form a platform for detection of cues and signaling and regulate the sensory capacity of cells and organisms among others by regulating the proper localization of signaling proteins in the cilium.
From studies in patient material and animal and cell models it is clear that the length and morphology of cilia are important for their function. Although the molecular mechanisms that regulate cilium length are poorly understood, it is clear that transport within cilia plays an important role. In previous studies we have identified novel proteins in C. elegans that regulate cilium length (Burghoorn et al., 2007 & 2010; Broekhuis et al., 2013; Van der Vaart et al., 2015) and extrapolated our findings to mammalian cells (Broekhuis et al., 2014; Kunova Bosakova et al., 2018 & 2019). We are currently using molecular genetic techniques, proteomics and imaging to identify and characterize novel proteins that regulate cilium length.
In addition, we contributed to several collaborative studies to characterize the polycyctins, involved in polycystic kidney disease and PACRG a structural ciliary protein (Kenter et al., 2019 & 2020; Loucks et al., 2016). We are currently using C. elegans to find out what determines where proteins localize within cilia, focusing on guanylate cyclases (GCYs) involved in NaCl detection in the environment. We found that some of these GCY proteins localize very specifically at the tip of the cilium (Van der Burght et al., 2020) and use genetics, proteomics, imaging and behavioral assay unravel the molecular mechanisms that control this.
Sensory signal transduction, focusing on salt (NaCl) taste
Despite its importance in our everyday life the molecular mechanisms of salt taste are not completely understood. We use C. elegans to identify proteins that are involved in NaCl taste and characterize their function. Important in these projects are behavioral assays, where we can quantify the preference of the animals for different NaCl concentrations.
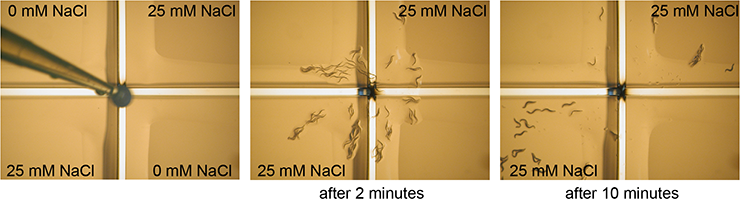
Adult C. elegans are transferred to a plate that is divided into four quadrants by plastic spacers. Two quadrants have been filled with agar containing 25 mM NaCl, two quadrants contain no NaCl. Just before the assay the plastic spacers have been covered with agar without NaCl. Animals go to the quadrants that they are attracted to. After 10 minutes the animals on each quadrant are counted, providing a measure for the response of the animals to NaCl.
We use behavioral, molecular genetic and cell biological techniques to identify and characterize novel proteins that play a role in NaCl taste. In addition, in collaboration with Prof. Dr. Ewout Hoorn we try to identify factors that contribute to the lower salt sensitivity of chronic kidney disease patients. To this end we are developing C. elegans that express the ENaC subunits involved in salt taste in humans, to further characterize the molecular mechanisms of salt taste.
In addition, we have developed a new paradigm to analyze behavioral plasticity. In this assay, worms learn to avoid NaCl, as they associate it with the absence of food. Using this assay and genetic tools several cells and genes involved in this behavior have been identified (Hukema et al., 2006 & 2008; Janssen et al., 2008 & 2009; Beets et al., 2012). Using in vivo Ca2+ imaging and computational modeling, we showed in collaboration with Dr Netta Cohen that the animals modulate the activity of the sensory neurons, allowing them to switch between attraction to and avoidance of NaCl, depending on their previous experience (Dekkers et al., 2021).
Maintenance of cell fate
Animals have many different cell types that each have a specific function. These functions are made possible by expression of specific genes, leading to the production of specific RNAs and proteins that together govern the structure and function of these cells: their cell fate. Research in the last decades has provided much information on the mechanisms that determine cell fate. However, during the life of the cell its fate should be maintained. This is especially important for long lived cells such as neurons.
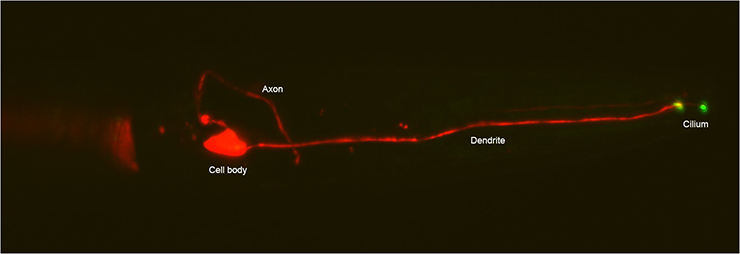
ASE neuron in head of an adult C. elegans marked with mCherry and showing GCY-22::GFP localizing at the base and tip of the cilium. Anterior is to the right.
We study maintenance of cell fate of the ASE salt sensing neurons in C. elegans. ASE neuron fate is determined by the terminal selector transcription factor CHE-1. How ASE cell fate is maintained was unclear, but it was thought to be mediated either by high levels or stability of CHE-1 mRNA or protein, or by chromatin modifications. Recently, in collaboration with Drs. Joleen Traets and Jeroen van Zon, we identified a novel mechanism that maintains cell fate over long timescales, despite molecular fluctuations that can lead to spontaneous loss of this differentiated state (Traets et al., 2021). Our simulations suggested that fluctuations in CHE-1 level are buffered by the reservoir of CHE-1 bound at its target promoters, which ensures continued che-1 expression by preferentially binding the che-1 promoter. Using CRISPR/Cas9 based genome editing and auxin mediated protein degradation we showed that che-1 expression was resilient to induced transient CHE-1 depletion, while both expression of CHE-1 targets and ASE function were lost. We identified a 130 bp che-1 promoter fragment responsible for this resilience. Deletion of a homeodomain binding site in this fragment caused stochastic loss of ASE identity long after its determination. Together, our results suggest that ASE cell fate is maintained by a mechanism, that relies on increased affinity of CHE-1 for its own promoter mediated by one of more homeodomain proteins binding to the che-1 promoter.
C. elegans as a model system
The nematode Caenorhabditis elegans is a well-established model organism that is often used to unravel basic processes at a molecular, cellular and organismal level. Important advantages of C. elegans are that they are easy to culture (at low costs), feed on bacteria, can be stored frozen, exist as hermaphrodites and males, have a generation time of 3 days, are transparent, small (1 mm) and have an invariant cell lineage which has been completely determined. In addition, many experimental tools are available, including genetic tools such as CRISPR/Cas9 mediated genome editing, transgenesis, RNAi and forward genetic screens, many imaging based approaches and various behavioral and organismal assays to study for example chemotaxis, locomotion, learning, memory and longevity.
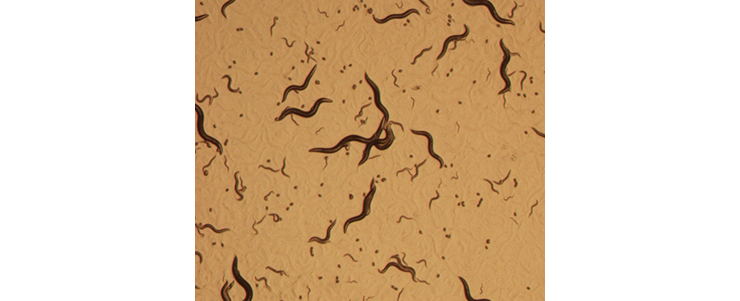
Culture of C. elegans on an agar plate with an E. coli lawn. Animals of different developmental stages (adult, larvae and eggs) can be seen, crawling through and leaving tracks in the E. coli lawn.
In most of the projects described earlier, we try to identify and characterize novel proteins involved in a process of interest. In addition, we use C. elegans to study proteins of interest, and how their functions are affected by mutations that have been found in human patients. In one project we study CaMKII, UNC-43 in C. elegans, in collaboration with Dr. Geeske van Woerden and Prof. Dr. Ype Elgersma. We are developing a model where we introduce mutations found in humans in one of the CaMKII genes into the C. elegans unc-43 gene. Using various behavioral assays, we analyze whether these mutations affect UNC-43 function.
In a second project, we use CRISPR/Cas9 to introduce the genes encoding human ENaC subunits (involved in NaCl taste) into the C. elegans genome, and express them in sensory neurons. If they are successfully expressed and functional, this will allow us to study how their function is affected by mutations and how human salt taste via ENaC is regulated.
Group members
Current group members
Manon Vleeming, PhD student
Laura van Vuuren, PhD student
Zoë de Wilt, Ba student Avans
Emma Boot, Msc Molecular Medicine student

Former group members (and their current whereabouts)
S. Rademakers, technician. Currently at Erasmus MC, Internal Medicine.
S. van der Burght, PhD student. Thesis: Molecular Mechanisms of Chemotaxis to Sodium Chloride in C. elegans. Currently at Gurdon Institute, Cambridge, UK.
A. Kenter, PhD student at Developmental Biology. Thesis: The Molecular Mechanisms of Polycystic kidney disease. Currently at Erasmus MC, Internal Medicine.
A. van der Vaart, post doc. Currently at the dept. of Biochemistry at Erasmus MC.
J.R. Broekhuis, PhD student. Thesis: Ciliary length control.
W.Y. Leong, PhD student.
O. Umuerri, PhD student. Thesis: Caenorhabditis elegans response to NaCl. Currently at Manitoba Health, Winnipeg, Canada.
E. Krpelanova, PhD student. Currently at World Health Organization.
M.P.J. Dekkers, PhD student. Thesis: Plasticity in Caenorhabditis elegans. Currently at Medartis, Basel, Switzerland.
R.K. Hukema, PhD student. Thesis: Gustatory behaviour in Caenorhabditis elegans. Currently at University of Applied Sciences, Rotterdam.
J. Burghoorn, PhD student. Thesis: Regulation of intraflagellar transport in the sensory cilia of Caenorhabditis elegans.
W.J. Lans, PhD student. Thesis: Making sense of G proteins: Genetic analysis of sensory G protein signalling in the nematode C. elegans. Currently at Dept. of Molecular Genetics, Erasmus MC.
Projects are available for Ba or MSc students.
Info: g.jansen@erasmusmc.nl
Publications
Scientific publications by the Jansen research group
2022
Chrystal, P.W., Lambacher, N.J., Doucette, L.P., Bellingham, J., Schiff, E.R., Noel, N.C.L., Li, C., Tsiropoulou, S., Casey, G.A., Zhai, Y., Nadolski, N.J., Majumder, M.H., Tagoe, J., D’Esposito, F., Cordeiro, F., Downes, S., Clayton-Smith, J., Ellingford, J., Genomics England Research Consortium, Mahroo, O.A., Hocking, J.C. Cheetham, M.E., Webster, A., Jansen, G., Blacque, O.E., Allison, W.T., Au, P-Y.B., MacDonald, I.M., Arno, G. & Leroux, M.R. (2022) The inner junction protein CFAP20 functions in motile and non-motile cilia and is critical for vision. Nat. Commun. 13, 6595. doi.org/10.1038/s41467-022-33820-w.
2021
Dekkers, M.P.J., Salfelder, F., Sanders, T., Umuerri, O., Cohen, N. & Jansen, G. (2021) Plasticity in gustatory and nociceptive neurons controls decision making in Caenorhabditis elegans salt navigation. Commun. Biol. 4: 1053. doi.org/10.1038/s42003-021-02561-9.
Traets, J.J.H., van der Burght, S.N., Rademakers, S., Jansen, G. & van Zon, J.S. (2021) Mechanism of life-long maintenance of neuron identity despite molecular fluctuations. eLife 10: e66955. doi: 10.7554/eLife.
2020
van der Burght, S., Rademakers, S., Johnson, J-L., Li, C., Kremers, G.J., Houtsmuller, A.B., Leroux, M.R. & Jansen, G. (2020) Ciliary tip signaling compartment is formed and maintained by intraflagellar transport. Curr. Biol. 30, 4299-4306. doi.org/10.1016/j.cub.2020.08.032.
Kenter, A.T., Rentmeester, E., van Riet, J., Boers, R., Boers, J., Ghazvini, M., Xavier, V.J., van Leenders, G.J.L.H., Verhagen, P.C.M.S., van Til, M.E., Eussen B., Losekoot, M., de Klein, A., Peters, D.J.M., van Ijcken, W.F.J., van de Werken, H.J.G., Zietse, R., Hoorn, E.J., Jansen, G., Gribnau, J.H. (2020) Cystic renal-epithelial derived Induced Pluripotent Stem Cells from Polycystic Kidney Disease patients. Stem Cells Transl. Med. 9, 478–490. doi: 10.1002/sctm.18-0283.
2019
Kenter, A.T., Van Rossum - Fikkert, S.E., Salih, M., Verhagen, P.C.M.S., Van Leenders, G.J.L.H., Demmers, J.A.A., Jansen, G., Gribnau, J.H., Zietse, R. & Hoorn, E.J. (2019) Identifying cystogenic paracrine signaling molecules in cyst fluid of patients with Polycystic Kidney Disease. Am. J. Physiol. Renal. Physiol. 316, F204-F213. doi: 10.1152/ajprenal.00470.2018.
Kunova Bosakova, M., Varecha, M., Fafilek, B., Barta, T., Gudernova, I., Balek, L., Potesil, D., Zieba, J.T., Song, J., Konik, P., Trantirek, L., Duran, I., Zdrahal, Z., Jansen, G., Fu, Z., Wan Ko, H., Hampl, A., Krakow, D., & Krejci, P. (2019) Fibroblast Growth Factor Signaling Controls Primary Cilia Length via Intestinal Cell Kinase. Proc. Natl. Acad. Sci USA. 116, 4316-4325. doi: 10.1073/pnas.1800338116.
2018
Kunova Bosakova, M., Varecha, M., Hampl, M., Duran, I., Nita, A., Buchtova, M., Dosedelova, H., Machat, R., Xie, Y., Ni, Z., Martin, J.H., Chen, L., Jansen, G., Krakow, D., & Krejci, P. (2018) Regulation of ciliary function by fibroblast growth factor signaling identifies FGFR3-related disorders achondroplasia and thanatophoric dysplasia as ciliopathies. Hum. Molec. Genet. 27, 1093-1105. doi: 10.1093/hmg/ddy031.
2016
Timbers, T., Garland, S.J., Mohan, S., Flibotte, S., Edgley, M., Muncaster, Q., Au, V., Li-Leger, E., Rosell, F., Cai, J., Rademakers, S., Jansen, G., Moerman, D.G. & Leroux, M.R. (2016) Accelerating gene discovery by phenotyping whole-genome sequenced multi-mutation strains and using the sequence kernel association test (SKAT). PLOS Genet. 12, e1006235. doi: 10.1371/journal.pgen.1006235.
Loucks, C.M., Bialas, N.J., Dekkers, M.P.J., Walker, D.S., Grundy, L.J., Li, C., Inglis, P.N., Kidad, K., William R. Schafer, W.R., Blacque, O.E., Jansen, G. & Leroux, M.R. (2016) PACRG, a protein linked to ciliary motility, mediates cellular signaling. Mol. Biol. Cell 27, 2133-2144. doi: 10.1091/mbc.E15-07-0490.
2015
Van der Vaart, A., Rademakers, S. & Jansen, G. (2015) DLK-1/p38 MAP Kinase Signaling Controls Cilium Length by Regulating RAB-5 Mediated Endocytosis in Caenorhabditis elegans. PLOS Genet. 11, e1005733. doi: 10.1371/journal.pgen.1005733.
González-Barrios, M., Fierro-González, J.C., Krpelanova, E., Mora-Lorca, J.A., Pedrajas, J.R., Peñate, X., Chavez, S., Swoboda, P., Jansen, G. & Miranda-Vizuete, A. (2015) Cis- and trans-regulatory mechanisms of gene expression in the ASJ sensory neuron of Caenorhabditis elegans. Genetics 200, 123-134. doi: 10.1534/genetics.115.176172.
2014
Broekhuis, J.R., Verhey, K.J. & Jansen, G. (2014) Regulation of cilium length and intraflagellar transport by the RCK-kinases ICK and MOK in renal epithelial cells. PLoS One 9, e108470. doi: 10.1371/journal.pone.0108470.
2013
Broekhuis, J.R., Rademakers, S., Burghoorn, J. & Jansen, G. (2013) SQL-1, homologue of the Golgi protein GMAP210, modulates Intraflagellar Transport in C. elegans. J. Cell Sci. 126, 1785-1795.doi: 10.1242/jcs.116640. IF 5.285.
Broekhuis, J.R., Leong, W.Y. & Jansen, G. (2013) Regulation of cilium length and intraflagellar transport. Int. Rev. Cell Mol. Biol. 303, 101-138. doi: 10.1016/B978-0-12-407697-6.00003-9.
Lans, H., Lindvall, J.M., Thijssen, K., Karambelas, A.E., Cupac, D., Fensgård, Ø., Jansen, G., Hoeijmakers, J.H.J., Nilsen, H., Vermeulen, W. (2013) DNA damage leads to progressive replicative decline but extends lifespan of long lived mutant animals. Cell Death Differ. 20, 1709-1718. doi: 10.1038/cdd.2013.126.
2012
Beets, I., Janssen, T., Meelkop, E., Temmerman, L., Suetens, N., Rademakers, S., Jansen, G., Schoofs, L. (2012) Vasopressin/oxytocin related signaling regulates gustatory associative learning in Caenorhabditis elegans. Science 338, 543-545. doi: 10.1126/science.1226860.
2010
Burghoorn, J., Dekkers, M.P.J., Rademakers, S., de Jong, T., Willemsen, R. Swoboda, P., & Jansen, G. (2010) Dauer pheromone and G protein signalling modulate the coordination of intraflagellar transport kinesin motor proteins in C. elegans. J. Cell Sci. 123, 2077-2084. doi: 10.1242/jcs.062885.
Lans, H., Marteijn J.A., Schumacher, B., Hoeijmakers, J.H.J., Jansen, G. & Vermeulen, W. (2010) Involvement of global genome repair, transcription-coupled repair and chromatin remodelling in UV DNA damage response changes during development. PLoS Genet. 6, e1000941. doi: 10.1371/journal.pgen.1000941.
2009
Janssen, T., Husson, S.J., Meelkop, E., Temmerman, L., Lindemans, M., Verstraelen, K., Rademakers, S., Mertens, I., Nitabach, M., Jansen, G. & Schoofs, L. (2009) Discovery and characterization of a conserved pigment dispersing factor-like neuropeptide pathway in Caenorhabditis elegans. J. Neurochem. 111, 228-241. doi: 10.1111/j.1471-4159.2009.06323.x.
Lans, H., Dekkers, M.P.J., Hukema, R.K., Bialas, N.J., Leroux, M.R. & Jansen, G. (2009) Signaling proteins that regulate NaCl chemotaxis responses modulate longevity in C. elegans. Ann. N. Y. Acad. Sci. 1170, 682-687. doi: 10.1111/j.1749-6632.2009.04362.x.
Luijsterburg, M.S., Dinant, C., Lans, H., Stap, J., Wiernasz, E., Lagerwerf, S., Warmerdam, D.O., Lindh, M., Brink, M.C., Dobrucki, J.W., Aten, J.A., Fousteri, M.I., Jansen, G., Dantuma, N.P., Vermeulen, W., Mullenders, L.H., Houtsmuller, A.B., Verschure, P.J. & van Driel, R. (2009) Heterochromatin protein 1 is recruited to various types of DNA damage. J. Cell Biol. 185, 577-586. doi: 10.1083/jcb.200810035.
2008
Hukema, R.K., Rademakers, S. & Jansen, G. (2008) Gustatory plasticity in C. elegans involves integration of negative cues and NaCl taste mediated by dopamine, serotonin, and glutamate. Learn. Mem. 15, 829-836. doi: 10.1101/lm.994408.
Janssen, T., Husson, S.J., Lindemans, M., Mertens, I., Rademakers, S., Ver Donck, K., Geysen, J., Jansen, G. & Schoofs, L. (2008) Functional characterization of three G protein-coupled receptors for pigment dispersing factors in Caenorhabditis elegans. J. Biol. Chem. 4283, 15241-15249. doi: 10.1074/jbc.M709060200.
Teng, M.S., Shadbolt, P., Fraser, A.G., Jansen, G. & McCafferty, J. (2008) Control of feeding behaviour in C. elegans by human G protein-coupled receptors permits screening of agonist-expressing bacteria. Proc. Natl. Acad. Sci. USA. 105, 14826–14831. doi: 10.1073/pnas.0803290105.
2007
Burghoorn, J., Dekkers, M.P.J., Rademakers, S., de Jong, T., Willemsen, R. & Jansen, G. (2007) Mutation of the MAP kinase DYF-5 affects docking and undocking of kinesin-2 motors and reduces their speed in the cilia of C. elegans. Proc. Natl. Acad. Sci. USA 104, 7157-7162. doi: 10.1073/pnas.0606974104.
Lans, H. & Jansen, G. (2007) Multiple sensory G proteins in the olfactory, gustatory and nociceptive neurons modulate longevity in Caenorhabditis elegans. Dev. Biol. 303, 474-482. doi: 10.1016/j.ydbio.2006.11.028.
2006
Hukema, R.K., Rademakers, S., Dekkers, M.P.J., Burghoorn, J. & Jansen, G. (2006) Antagonistic sensory cues generate gustatory plasticity in Caenorhabditis elegans. EMBO J. 25, 312-322. doi: 10.1038/sj.emboj.7600940.
Jansen, G. & Ségalat, L. (2006) Behavioral genetics in the nematode Caenorhabditis elegans. In ‘Neurobehavioral Genetics: Methods and Applications’, BC Jones & P Mormede Eds. CRC Press.
Lans, H. & Jansen, G. (2006) Non-cell and cell-autonomous G protein-signaling converges with Ca2+/mitogen-activated protein kinase signaling to regulate str-2 receptor gene expression in C. elegans. Genetics 173, 1-13. doi: 10.1534/genetics.106.058750.
Teng, M.S., Dekkers, M.P.J., Ng, B.L., Rademakers, S., Jansen, G., Fraser, A.G. & McCafferty, J. (2006) Expression of mammalian GPCRs in C. elegans generates novel behaioural responses to human ligands. BMC Biology 4, 22. doi: 10.1186/1741-7007-4-22.
2005
Pellis-van Berkel, W., Verheijen, M.H.G., Cuppen, E., Asahina, M., de Rooij, J., Jansen, G., Plasterk, R.H.A., Bos, J.L. & Zwartkruis, F.J.T. (2005) Requirement of the C. elegans RapGEF pxf-1 and rap-1 for epithelial integrity. Mol. Biol. Cell 16, 106-116. doi: 10.1091/mbc.e04-06-0492.
2004
Fukuto, H.S., Ferkey, D.M., Apicella, A.J., Lans, H., Sharmeen, T., Chen, W., Lefkowitz, R.J., Jansen, G., Schafer, W.R. & Hart, A.C. (2004) G protein-coupled receptor kinase function is essential for chemosensation in C. elegans. Neuron 42, 581-593. doi: 10.1016/s0896-6273(04)00252-1.
Lans, H., Rademakers, S. & Jansen, G. (2004) A Network of Stimulatory and Inhibitory G Subunits Regulates Olfaction in Caenorhabditis elegans. Genetics 167, 1677-1687. doi: 10.1534/genetics.103.024786. IF 4.562.
2003
Cuppen, E., van der Linden, A.M., Jansen, G. & Plasterk, R.H.A. (2003) Proteins interacting with Caenorhabditis elegans G subunits. Comp. Funct. Genom. 4, 479-491. doi: 10.1002/cfg.318.
2002
Jansen, G. (2002) Gene inactivation in Caenorhabditis elegans. Curr. Genomics 3, 2002, 59-67.
Jansen, G., Weinkove, D. & Plasterk, R.H.A. (2002) The G-protein gamma subunit gpc-1 of the nematode C. elegans is involved in taste adaptation. EMBO J. 21, 986-994. doi: 10.1093/emboj/21.5.986.
2001
De Voer, G., Jansen, G., van Ommen, G.J., Peters, D.J. & Taschner, P.E. (2001) Caenorhabditis elegans homologues of the CLN3 gene, mutated in juvenile neuronal ceroid lipofuscinosis. Eur. J. Paediatr. Neurol. 5 Suppl A:115-120. doi: 10.1053/ejpn.2000.0446.
Teaching activities
Program director of the MSc program Molecular Medicine
(link to https://www.eur.nl/en/research-master/molecular-medicine)
Coordinator minors Erasmus MC
(link to https://www.eur.nl/erasmusmc/onderwijs/minor)
Leader of the project to restructure the Erasmus MC minors
Member of the Curriculum Committee ErasmusArts 2030
(link to Bachelor Geneeskunde | Erasmus MC | Erasmus University Rotterdam)
External links
Cildb knowledgebase http://cildb.i2bc.paris-saclay.fr/
Syscilia http://syscilia.org/index.shtml
CiliaCarta https://tbb.bio.uu.nl/john/syscilia/ciliacarta/
Wormbase https://wormbase.org/
Wormbook http://www.wormbook.org/
Wormatlas https://www.wormatlas.org/
