About Dr. E. (Eskeatnaf) Mulugeta
Introduction
Systems biology of signaling and transcription factors (SBSTF)
Eskeatnaf Mulugeta has a multi-disciplinary background and diverse research interest. His current research applies cutting-edge computational and molecular biology techniques to understand the systems biology of signaling and transcription factor networks during development and disease.
Research
Cellular development and differentiation is a dynamic and tightly controlled process that requires the cells ability to adjust to new situation by changing its phenotype and functioning. This complex process is orchestrated by transcriptional regulation of genes that involve serval layers of regulatory modules. Signaling pathways and transcription factors (TFs) are important components of these gene regulatory modules. Cells sense the change in their environment and transmit the message to the nucleus using signal transduction pathways. In the nucleus, the message is interpreted by a coordinated action of signal transduction proteins and TFs to modulate the expression level of target genes by changing the 3D genome confirmation, chromatin environment (histone and DNA modifications), and accessibility of genes to the transcription machinery (Figure 1). This process of cellular information reception, transduction, and interpretation is strictly controlled at every level involving several genes that are organized in different gene regulatory networks (biological circuits). The cooperative action of these gene regulatory networks provide efficient information flow and interpretation and also stop when it is not needed. Mutations in components of cellular information flow and interpretation system or changes in the process result in developmental abnormalities and cause chronic diseases, including cancer.
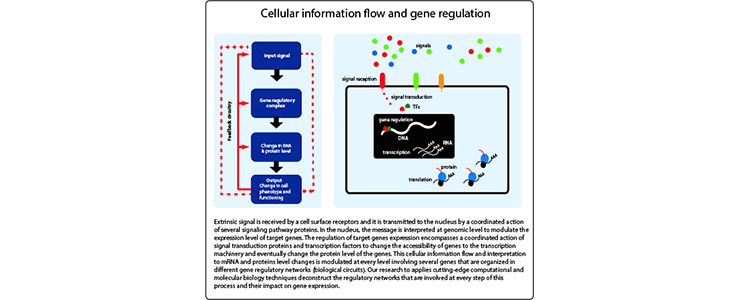
Traditionally, and also due to limitations of the available techniques, these type of complex and dynamic cellular systems have been studied by a reductionist approach, looking at single genes or a limited number of genes. Recent advances in gene editing technologies, single-cell sequencing and other high-throughput methods are opening a new avenue in how such system biology problems could be approached in a holistic way. My research uses a holistic approach applying existing/emerging state of the art computational and molecular biology techniques and also by developing new approaches to unravel the systems biology of signaling pathways and transcription factors. My main objective is to deconstruct the regulatory network (biological circuits) of signaling pathways and transcription factors (TFs) during development and disease by using pluripotent mouse embryonic stem cells and their neural and hematopoietic differentiation as a model. This includes :
- Understanding the dynamics of signaling pathways and TFs
- Elucidating the cross-talk between signaling pathways and TFs
- Elucidating the crosstalk between different signaling pathways
- Deciphering the hierarchy of signaling pathways and TFs
- Understanding the eventual impact on 3D genome organization, chromatin, and transcription.
The techniques we use (will use) include:
- Embryonic stem cell culture and differentiation
- Crisper-cas9 based massively parallel gene perturbation
- Genomic:
* RNA sequencing
* Methylome: Reduced representation bisulfite sequencing (RRBS)
* Chromatin accessibility (ATAC-seq)
* Immunoprecipitation with massively parallel DNA sequencing (ChIP-seq)
* A family of methods for analyzing chromatin interactions
(Hi-C/3C-Seq/Capture-C,T2C) - Single cell Genomics:
* Single cell RNA-seq,
* Single cell ATAC-seq,
* Single cell Methylome - In addition, we develop novel approaches
Other project: naked mole rat
Naked mole rat: a model for the study of mammalian eusociality, longevity & cancer resistance
The naked mole rat (NMR) (Heterocephalus glaber) is a subterranean rodent endemic to parts of East Africa that has become famous for several unique characteristics such as an extraordinarily long life, cancer resistance, and eusociality. The NMR lives for more than 30 years; 6-7 times longer than that of similarly sized house mouse and 5 times longer than expected based on its body size. This longevity of NMR is also accompanied by a good health maintaining normal activity, body composition, reproductive and physiological functions for the majority of its lifespan. The NMR is one of the only two known mammals with eusocial organisation (the highest level of organization of animal sociality), similar to that of bees and ants. The NMRs live in a large colony od around 300 individuals with very few breeding animals (one breeding female, “queen” and up to 3 breeding males) where the other sexually supressed members of the colony perform activities for the well-being of the society, caring for the pups, foraging for food, and defending the colony. In collaboration with Edith Heard (Institut Curie, Paris), Elizabeth Blackburn (University of California, San Francisco), and Chris Faulkes (Queen Mary University of London), we are applying genomic and epigenomic techniques to gain new insights into the extraordinary characteristics of naked mole rats: eusociality, longevity and cancer resistance.

Education and career
| Name | Eskeatnaf Mulugeta |
| e.mulugeta@erasmusmc.nl, eskeww@gmail.com | |
| Unique identifiers | ORCiD: 0000-0003-4045-4835 |
| ResearcherID: H-2640-2016 | |
| Twitter:@eskeww | |
| Linkedin:https://goo.gl/F4lgqQ | |
| Google scholar:https://goo.gl/QgNEz9 |
Education
| 2008–2012 | Doctor of Philosophy (PhD) Dept. of Reproduction and Development, Erasmus University Rotterdam, Erasmus University Medical Center (Erasmus MC), Rotterdam, The Netherlands |
| 2007–2009 | Master Molecular Medicine, Erasmus University Rotterdam, Erasmus MC |
| 2004–2006 | Master Bioinformatics, Wageningen University |
| 2002–2004 | Master Biotechnology, Wageningen University |
| 1996–2001 | Bachelor of Pharmacy (BPharm), Addis Ababa University |
Research experience
| 2018–present | Junior group leader, Dept. of Cell Biology, Erasmus MC, Rotterdam, The Netherlands. |
| 2013 - 2017 | Postdoctoral researcher, Institute Curie, Paris, France. Prof. Edith Heard |
| 2012 - 2013 | Scientific researcher, Dept. of Reproduction and Development, Erasmus MC, Rotterdam (The Netherlands). Prof. Joost Gribnau |
| 2008–2012 | PhD student, Dept. of Reproduction and Development, Erasmus University Rotterdam, Erasmus MC, Rotterdam, The Netherlands. Prof. Joost Gribnau |
Publications
Peifen Zhang, Paula Haro Angles, Hong Zhang, Tingyue Li, Roger Mulet-Lazaro, Wilfred F.J. van IJcken, Sjaak Philipsen, Ruud Delwel, Joost Gribnau, Danny Huylebroeck, Frank Grosveld, Claire Rougeulle, Eskeatnaf Mulugeta. A Novel XACT lncRNA Transcript with Functions Transcending X-Chromosome Inactivation, bioRxiv 2025.01.03.631201; doi: https://doi.org/10.1101/2025.01.03.631201
Ruizhi Deng, Elena Perenthaler, Anita Nikoncuk, Soheil Yousefi, Kristina Lanko, Rachel Schot, Michela Maresca, Michael J. Parker, Wilfred F.J. van Ijcken, Joohyun Park, Marc Sturm, Tobias B. Haack, Genomics England Research Consortium, Gennady V Roshchupkin, Eskeatnaf Mulugeta, Tahsin Stefan Barakat. A novel functional genomics atlas coupled with convolutional neural networks facilitates clinical interpretation of disease relevant variants in non-coding regulatory elements. Medrxiv, April 2024, DOI: https://doi.org/10.1101/2024.04.13.24305761
Lucas Kuijpers, Bastian Hornung, Mirjam CGN van den Hout-van Vroonhoven, Wilfred FJ van IJcken, Frank Grosveld, Eskeatnaf Mulugeta. Split Pool Ligation-based Single-cell Transcriptome sequencing (SPLiT-seq) data processing pipeline comparison. BMC Genomics, 2024. https://doi.org/10.1186/s12864-024-10285-3
Lucie Kulhankova, Eric Bindels, Manfred Kayser, Eskeatnaf Mulugeta. Deconvoluting multi-person biological mixtures and accurate characterization and identification of separated contributors using non-targeted single-cell DNA sequencing. Forensic Science International: Genetics, 2024. https://doi.org/10.1016/j.fsigen.2024.103030
Yuvia A Pérez-Rico, Aurélie Bousard, Lenka Henao Misikova, Eskeatnaf Mulugeta, Sergio F de Almeida, Alysson R Muotri, Edith Heard, Anne-Valerie Gendrel. Transcriptional perturbation of LINE-1 elements reveals their cis-regulatory potential. bioRxiv, 2024. https://doi.org/10.1101/2024.02.20.581275
Ruben Boers, Joachim Boers, Beatrice Tan, Marieke E. van Leeuwen, Evelyne Wassenaar, Erlantz Gonzalez Sanchez, Esther Sleddens, Yasha Tenhagen, Eskeatnaf Mulugeta, Joop Laven, Menno Creyghton, Willy Baarends, Wilfred F. J. van IJcken & Joost Gribnau. Retrospective analysis of enhancer activity and transcriptome history. Nat Biotechnol (2023). https://doi.org/10.1038/s41587-023-01683-1
Lucie Kulhankova, Diego Montiel Gonzáles, Eric Bindels, Daniel Kling, Manfred Kayser #, Eskeatnaf Mulugeta # Single-cell transcriptome sequencing allows genetic separation, characterization and identification of individuals in multi-person biological mixtures. Commun Biol 6, 201 (2023). https://doi.org/10.1038/s42003-023-04557-z
Lucile Marion-Poll, Jean-Pierre Roussarie, Lieng Taing, Cloelia Dard-Dascot, Nicolas Servant, Yan Jaszczyszyn, Emmanuelle Jordi, Eskeatnaf Mulugeta, Denis Hervé, Déborah Bourc’his, Paul Greengard, Claude Thermes & Jean-Antoine Girault. DNA methylation and hydroxymethylation characterize the identity of D1 and D2 striatal projection neurons. Communications Biology volume 5, Article number: 1321 (2022). DOI 10.1038/s42003-022-04269-w
Ruben Boers, Joachim Boers, Beatrice Tan, Evelyne Wassenaar, Erlantz Gonzalez Sanchez, Esther Sleddens, Yasha Tenhagen, Marieke E. van Leeuwen, Eskeatnaf Mulugeta, Joop Laven, Menno Creyghton, Willy Baarends, Wilfred F. J. van IJcken, Joost Gribnau. Retrospective analysis of enhancer activity and transcriptome history. Preprint 2021: doi: https://doi.org/10.1101/2021.09.07.459233
Soheil Yousefi, Ruizhi Deng, Kristina Lanko, Eva Medico Salsench, Anita Nikoncuk, Herma C. van der Linde, Elena Perenthaler, Tjakko J. van Ham, Eskeatnaf Mulugeta* & Tahsin Stefan Barakat*. Comprehensive multi-omics integration identifies differentially active enhancers during human brain development with clinical relevance. (# corresponding authors) Genome Med 13, 162 (2021). https://doi.org/10.1186/s13073-021-00980-1
Willeke de Haan, Wouter Dheedene, Katerina Apelt, Sofiane Décombas-Deschamps, Stefan Vinckier, Stefaan Verhulst, Andrea Conidi, Thomas Deffieux, Michael W Staring, Petra Vandervoort, Ellen Caluwé, Marleen Lox, Inge Mannaerts, Tsuyoshi Takagi, Joris Jaekers, Geert Berx, Jody Haigh, Baki Topal, An Zwijsen, Yujiro Higashi, Leo A van Grunsven, Wilfred F J van IJcken, Eskeatnaf Mulugeta, Mickael Tanter, Franck P G Lebrin, Danny Huylebroeck, Aernout Luttun. Endothelial Zeb2 preserves the hepatic angioarchitecture and protects against liver fibrosis. Cardiovasc Research, 2021, doi:10.1093/cvr/cvab148
Lisanne Groeneveldt, Tim Herpelinck, Marina Maréchal, Constantinus Politis, Wilfred van IJcken, Danny Huylebroeck, Liesbet Geris, Eskeatnaf Mulugeta and Frank Luyten. The bone-forming properties of periosteum-derived cells differ between harvest sites. Front. Cell Dev. Biol., 25 November 2020 | https://doi.org/10.3389/fcell.2020.554984
Miyata M, Gillemans N, Hockman D, Demmers JA, Cheng JF, Hou J, Salminen M, Fisher C, Taylor S, Gibbons RJ, Ganis JJ, Zon LI, Grosveld F, Mulugeta E, Sauka-Spengler T, Higgs DR, Philipsen S. An evolutionary ancient mechanism for regulation of hemoglobin expression in vertebrate red cells. Blood. 2020 May 12:blood.2020004826. doi: 10.1182/blood.2020004826
Parenti I, Diab F, Gil SR, Mulugeta E, Casa V, Berutti R, Brouwer RWW, Dupé V, Eckhold J, Graf E, Puisac B, Ramos F, Schwarzmayr T, Gines MM, van Staveren T, van IJcken WFJ, Strom TM, Pié J, Watrin E, Kaiser FJ, Wendt KS. MAU2 and NIPBL Variants Impair the Heterodimerization of the Cohesin Loader Subunits and Cause Cornelia de Lange Syndrome. Cell Rep. 2020 May 19;31(7):107647 https://doi.org/10.1016/j.celrep.2020.107647
Ruben Dries, Agata Stryjewska, Kathleen Coddens, Satoshi Okawa, Tineke Notelaers, Judith Birkhoff, Mike Dekker, Catherine M Verfaillie, Antonio Del Sol, Eskeatnaf Mulugeta, Andrea Conidi, Frank G Grosveld, Danny Huylebroeck. Integrative and perturbation based analysis of the transcriptional dynamics of TGFβ/BMP system components in transition from embryonic stem cells to neural progenitors, Stem Cells 2019, DOI https://doi.org/10.1002/stem.3111
Isabelle Blomfield, Brenda Rocamonde, Maria Del Mar Masdeu, Eskeatnaf Mulugeta, Stefania Vaga, Debbie LC van den Berg, Emmanuelle Huillard, Noelia Urbán, Francois Guillemot. Id4 eliminates the pro-activation factor Ascl1 to maintain quiescence of adult hippocampal stem cells. (Elife 2019, DOI: 10.7554/eLife.48561, Biorxiv 2018, doi: https://doi.org/10.1101/426015)
Josef Redolfi, Yinxiu Zhan, Christian Valdes-Quezada, Mariya Kryzhanovska, Isabel Guerreiro, Vytautas Iesmantavicius, Tim Pollex, Ralph S Grand, Eskeatnaf Mulugeta, Jop Kind, Guido Tiana, Sebastien A Smallwood, Wouter de Laat, Luca Giorgetti. DamC reveals principles of chromatin folding in vivo without crosslinking and ligation. Nature Structural & Molecular Biology, volume 26, pages471–480 (2019), doi: https://doi.org/10.1038/s41594-019-0231-0
Ilaria Parenti, Farah Diab, Sara Ruiz Gil, Eskeatnaf Mulugeta, Valentina Casa, Riccardo Berutti, Rutger WW Brouwer, Valerie Dupe, Juliane Eckhold, Elisabeth Graf, Beatriz Puisac, Feliciano Ramos, Thomas Schwarzmayr, Thomas van Staveren, Wilfred FJ van IJcken, Tim M Strom, Juan Pie, Erwan Watrin, Frank J Kaiser, Kerstin S Wendt. MAU2 and NIPBL variants in Cornelia de Lange syndrome reveal MAU2-independent loading of cohesin and uncover a protective mechanism against early truncating mutations in NIPBL. 2018 Biorxiv, preprint available at: https://www.biorxiv.org/content/10.1101/477752v1
Eskeatnaf Mulugeta*, Lucile Marion-Poll, David Gentien, Elizabeth Helen Blackburn, Chris G. Faulkes*, Edith Heard. Unravelling the pathways underlying naked mole-rats eusociality. * Corresponding authors. 2018 Biorxiv, preprint available at: https://goo.gl/d3e6XB
Eskeatnaf Mulugeta, Evelyne Wassenaar, Esther Sleddens-Linkels, Wilfred FJ van IJcken, Edith Heard, J. Anton Grootegoed, Walter Just, Joost Gribnau, and Willy M Baarends (2106) Genomes of Ellobius species provide insight into the evolutionary dynamics of mammalian sex chromosomes. Genome Research 2016, 26(9):1202-10.
Marie Schoumacher, Stéphanie Le Corre, Alexandre Houy, Eskeatnaf Mulugeta, Marc-Henri Stern, Sergio Roman-Roman, Raphaël Margueron (2016) Uveal melanoma cells are resistant to EZH2 inhibition regardless of BAP1 status. Nature Medicine 22: 577–78.
Anthony Ferrari, Anne Vincent-Salomon, Xavier Pivot, Anne-Sophie Sertier, Emilie Thomas, Laurie Tonon, Sandrine Boyault, Eskeatnaf Mulugeta, Isabelle Treilleux, Gaëtan MacGrogan, Laurent Arnould, Janice Kielbassa, Vincent Le Texier, Hélène Blanché, Jean-François Deleuze, Jocelyne Jacquemier, Marie-Christine Mathieu, Frédérique Penault-Llorca, Frédéric Bibeau, Odette Mariani,Cécile Mannina, Jean-Yves Pierga, Olivier Trédan, Thomas Bachelot, Hervé Bonnefoi, Gilles Romieu, Pierre Fumoleau, Suzette Delaloge, Maria Rios, Jean-Marc Ferrero, Carole Tarpin, Catherine Bouteille, Fabien Calvo, Ivo Glynne Gut, Marta Gut, Sancha Martin, Serena Nik-Zainal, Michael R. Stratton, Iris Pauporté, Pierre Saintigny, Daniel Birnbaum, Alain Viari and Gilles Thomas (2016) A whole-genome sequence and transcriptome perspective on HER2-positive breast cancers. Nature Communications 2016, 7: 12222.
Federica Federici*, Eskeatnaf Mulugeta*, Sam Schoenmakers, Evelyne Wassenaar, Jos Hoogerbrugge, G. W. van der Heijden, Wiggert van Cappellen, Johan Slotman, Wilfred IJcken, Joop Laven, Anton Grootegoed, Willy Baarends (2015) Incomplete meiotic sex chromosome inactivation in the domestic dog. BMC Genomics 2015, 16:291. (* equal contribution)
Cristina Gontan, Eskeatnaf Mulugeta, Tahsin Stefan Barakat, Eveline Rentmeester, Wilfred van IJcken, Jeroen Demmers, Joost Gribnau, Anton Grootegoed (2012) RNF12 initiates X chromosome inactivation by targeting REX1 for degradation. Nature 2012, 485:386-90.
Tahsin Stefan Barakat, Nilhan Gunhanlar, Cristina Gontan Pardo, Eskeatnaf Mulugeta, Mehrnaz Ghazvini, Ruben Boers, Annegien Kenter, Eveline Rentmeester, J Anton Grootegoed, Joost Gribnau (2011) RNF12 activates Xist and is essential for X chromosome inactivation. PLoS Genetics 2011, 7(1):e1002001.
Eskeatnaf Mulugeta, Willy M Baarends, Joost Gribnau, Anton Grootegoed (2010) Evaluating the relationship between spermatogenic silencing of the X chromosome and evolution of the Y chromosome in chimpanzee and human. PLoS One 2010, 5(12):e15598.
Eskeatnaf Mulugeta, Evelyne Wassenaar, Jos W Hoogerbrugge, Esther Sleddens-Linkels, Marja Ooms, Zu-Wen Sun, Wilfred FJ van Ijcken, Anton Grootegoed, Willy M Baarends (2010) The ubiquitin-conjugating enzyme HR6B is required for maintenance of X chromosome silencing in mouse spermatocytes and spermatids. BMC Genomics 2010, 11:367.
Iris Jonkers, Tahsin Stefan Barakat, Eskeatnaf Mulugeta, Kim Monkhorst, Annegien Kenter, Eveline Rentmeester, Frank Grosveld, Anton Grootegoed, Joost Gribnau (2009) RNF12 is an X-encoded dose-dependent activator of X chromosome inactivation. Cell 2009, 139(5):999-1011.
Kim Monkhorst, Bas de Hoon, Iris Jonkers, Eskeatnaf Mulugeta, Wouter Monkhorst, Jos Hoogerbrugge, Eveline Rentmeester, Hans V Westerhoff, Frank Grosveld, Anton Grootegoed, Joost Gribnau (2009) The probability to initiate X chromosome inactivation is determined by the X to autosomal ratio and X chromosome specific allelic properties. PLoS One 2009, 4(5): e5616
Wessel N van Wieringen, Jeroen A M Belien, Sjoerd J Vosse, Eskeatnaf Mulugeta, Bauke Ylstra (2006) ACE-it: a tool for genome-wide integration of gene dosage and RNA expression data. Bioinformatics 2006, 22(15):1919-20.
Jacco van Rheenen, Eskeatnaf Mulugeta, Hans Janssen, Jero Calafat, Kees Jalink (2005) PIP2 signaling in lipid domains: a critical re-evaluation. The EMBO Journal 2005, 4;24(9):1664-73.
Eskeatnaf Mulugeta. Dynamics of mammalian X and Y chromosomes in evolution and development (ISBN:978-94-6182-131-7)
Positions
We have several master projects available.
Please contact e.mulugeta@erasmusmc.nl
We are looking for students with strong interest in:
- Dry lab experiments (bio-informatics and computational biology)
- Wet lab experimental work that involves:
- stem cell differentiation in cell culture (mESC and induced pluripotent cells)
- gene perturbation experiments (via CRISPR/Cas)
- single cell RNA and DNA sequencing
Group members
Peifen Zhang, PhD student
Hong Zhang, PhD student
Lab alumni
Iris Bakker (Bachelor Nanobiology, TU Delft)
Caroline Bastiaanssen (Master Nanobiology, TU Delft)
Thomas Boerstra (former PhD student, Erasmus MC)
Pierangela Chiafele (Master Molecular Medicine, Erasmus MC)
Erlantz Gonzalez (Master Molecular Medicine, Erasmus MC)
Meike Pollemans, MSc student Bio-Pharmaceutical Sciences at Leiden University)
Júlia Sansa (Universitat de Barcelona, Erasmus+ student)
