About Dr. R.A. (Raymond) Poot
Introduction
Role of transcription factors and chromatin modifiers in establishing normal cognitive ability
Group leader: Raymond Poot
Group members:
Eline Koornstra (PhD student)
Eva van Zanten (PhD student)
Introduction
Gene transcription is regulated by sequence-specific transcription factors and chromatin modifiers. My group aims to address questions such as; 1) How do transcription factors and chromatin modifiers at enhancers and promoters regulate transcription? 2) Why and how do mutations in chromatin modifiers and transcription factors often cause cognitive disabilities, such as autism.
We try to answer these questions using human neural stem cells and zebrafish as model systems. We previously showed that Sox2 (mutated in patients with SOX2 anophtalmia syndrome) interacts with chromatin remodeling factor Chd7 (mutated in patients with CHARGE syndrome) and together they regulate a set of common target genes. Intriguingly, SOX2 syndrome and CHARGE syndrome have overlapping features that are also observed in patients that have mutations in some of the Sox2-Chd7 target genes (see Figure 1 and Engelen et al. 2011 for details).

Figure 1
(legend) Hypothetical model for mechanistic links between shared malformations in different human syndromes. Syndromes are in gray lettering, haploinsufficiency for the gene associated with the syndrome is in black lettering, associated malformations or defects are indicated by black arrows, transcriptional regulation of genes by Sox2+Chd7 in NSCs are indicated by dashed red arrows and shared malformations or defects are indicated in blue. (Details see Engelen et al. 2011) (https://www.ncbi.nlm.nih.gov/pubmed/21532573)
We set-up a method to ChIP DNA marked by certain histone modifications and identify the proteins bound to these types of chromatin (ChIP-MS) and used this to identify transcription and chromatin factors that bind to highly active enhancers and promoters (see Figure 2 and Engelen, Brandsma et al. 2015 for details).
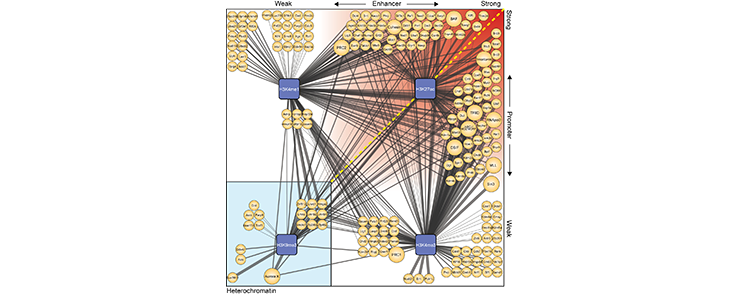
Figure 2
(legend) ChIP-MS predicts protein factors at active enhancers, active promoters and heterochromatin. (Details see Engelen, Brandsma et al. 2015) (https://www.ncbi.nlm.nih.gov/pubmed/25990348)
Recently it has emerged that chromatin modifier genes represent nearly half of the high confidence autism genes (https://gene.sfari.org/database/human-gene/). This high enrichment has sparked our interest to understand how mutations in chromatin modifier genes cause autism. We have purified transcription factors from neural stem cells and identified their interaction partners by mass spectroscopy. The resulting protein interaction network is enriched for proteins whose genes are mutated in patients with autism or schizophrenia (see Figure 3, and Moen et al 2017 for details). Interestingly, mutations in network protein genes are often loss-of-function in patients with autism with low IQ, but harbour missense mutations in patients with autism and normal IQ or patients with schizophrenia, suggesting that the severity of disruption of the same protein network correlates with the severity of the cognitive disability (see Moen et al 2017 for details).
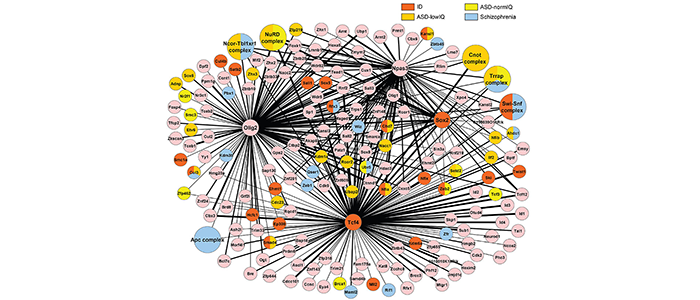
Figure 3
(legend) Protein interaction network in neural stem cells. Interaction network represents proteins present in two or more purifications of FLAG-tagged Tcf4, Olig2, Npas3 or Sox2 from neural stem cells. Protein complexes are larger circles, thickness of the edges (black lines) gives an indication of protein quantity in the purified samples. Colouring according to protein/gene mutated in patients with ID, ASD-normIQ, ASD-lowIQ or schizophrenia. ID; intellectual disability, ASD-normIQ; autism with normal IQ (>90). ASD-lowIQ; autism with low IQ (≤90).
(details see Moen et al., 2017) (https://www.ncbi.nlm.nih.gov/pubmed/28375211)
We use human neural stem cells with mutations in autism-related chromatin modifiers and purification of autism-related chromatin modifiers from human neural stem cells to better understand what these chromatin modifiers do in human neural stem cells and what molecular pathways are affected when they are mutated. Aberrant human neural stem cell proliferation is implied in early post-natal excess brain growth and the resulting temporal or permanent macrocephaly associated with autism, making human neural stem cells an excellent model system to identify autism-related molecular pathways.

(legend) Zebrafish
We have recently started working with zebrafish to address how mutations in autism-related chromatin modifiers lead to autism-relevant phenotypes. Zebrafish are transparent early in development allowing a unique view on early brain development and its aberrations upon autism mutations.
Report Open Science and DORA
Publications
Selected publications:
1. Meert L., Pelicano de Almeida M., Dekker M.R., Dekkers D.H.W., Nowosad K., Huylebroeck D., van den Hout M., Ozgür Z., van IJcken W.F.J., Demmers J., Fornerod M., Poot R.A. (2025) A CHD8-TRRAP axis facilitates MYC and E2F target gene regulation in human neural stem cells
iScience 28:111978.
2. Knol M.J.*, Poot R.A.*, et al. (2024) Genetic variants for head size share genes and pathways with cancer.
Cell Rep Medicine 5:101529
3. Quevedo M., Meert L., Dekker M.R., Dekkers D.H.W., Brandsma J.H., van den Berg D.L.C., Ozgür Z., IJcken W.F.J.V., Demmers J., Fornerod M., Poot R.A. (2019) Mediator complex interaction partners organize the transcriptional network that defines neural stem cells.
Nature Communications 10:2669.
4. Engelen E., Brandsma J.H., Moen M.J., Signorile L., Dekkers D.H., Demmers J., Kockx C.E., Ozgür Z., van IJcken W.F., van den Berg D.L., Poot R.A. (2015) Proteins that bind regulatory regions identified by histone modification chromatin immunoprecipitations and mass spectrometry.
Nature Communications 6:7155.
5. Engelen E., Akinci U., Bryne J.C., Hou J., Gontan C., Moen M., Szumska D., Kockx C., van IJcken W., Dekkers D.H.W, Demmers J., Rijkers E.J., Bhattacharya S., Philipsen S., Pevny L.H., Grosveld F.G., Rottier R.J., Lenhard B., Poot R.A. (2011) Sox2 cooperates with Chd7 to regulate genes that are mutated in genetically unrelated human syndromes.
Nature Genetics 43: 607-611
6. Van den Berg D.L. Snoek, T., Mullin N., Yates, A., Bestarosti, K., Demmers, J., Chambers I., Poot R.A. (2010) An Oct4-centered interaction network in embryonic stem cells.
Cell Stem Cell 6: 369-381
Student projects
Research projects for BSc and MSc students from Molecular Medicine, Nanobiology and other programs involve:
- CRISPR/Cas9-mediated genome editing to introduce mutations, affinity tags and reporters in human ES cells, neural stem cells and zebrafish
- Neuronal differentiation of human neural stem cells and human ES cells
- ChIP-Seq to explore binding sites of chromatin modifiers and histone modifications in human neural stem cells
- Protein purification from human neural stem cells to hunt for interaction partners of autism-related chromatin modifiers
- Zebrafish microscopy and behavioral studies
When you perform a research project in our laboratory you can expect:
- Daily supervision, biweekly work discussions and weekly plenary departmental project discussions
- Frequent seminars by national and international speakers
- Well-equipped laboratories with state-of-the-art imaging, genomics and proteomics facilities
- A critical but supportive research environment
- Freedom to pursue original ideas
Contact address: r.poot@erasmusmc.nl
Investeer in wetenschap voor de toekomst van Nederland
Vrij-wetenschappelijk-onderzoek-verdient-meer-investeringen-minEZK
Vrij-wetenschappelijk-onderzoek-verdient-meer-investeringen-minOCW
29 oktober 2020
Investeer in wetenschap voor de toekomst van Nederland
VK - Veel meer investeren in wetenschap is mogelijk en noodzakelijk voor de BV Nederland
Alumni
Postdocs
Katalin Tacs – Scientist at MRC London Institute of Medical Sciences
Umut Akinci – Scientist at Janssen Pharmaceutical Companies of Johnson&Johnson
PhD students
Debbie van den Berg – Group leader at Department of Cell Biology, Erasmus MC
Erik Engelen – Principal Scientist at Roche Diagnostics, Germany
Johan Brandsma – Informatics expert at Dutch Ministry of Internal Affairs
Maaike Moen – Biomolecular Scientist / Advisor at National Healthcare Institute
Marti Quevedo – Post Doc at Umea Plant Science Centre, Sweden
Lize Meert - program manager at ZonMW, the Netherlands
Mariana Pelicano d'Almeida - postdoctoral researcher at University of Porto, Portugal
Master students
Tim Snoek – Project coordinator at Springer Nature Publishers
Chantal Achterberg – Data Analyst at CGI, Olympic silver and bronze medallist
Sophia Karkampouna – Post Doc at University of Bern
Luca Signorile – Senior Computational Chemist at SOM Biotech, Barcelona
Hieab Adams – Group leader at Department of Epidemiology, Erasmus MC
Isabel Gordaliza – PhD student at IRB Barcelona
Adela Lleches – Quality Assurance Technician at Sanofi, Barcelona
Eva Niggl – Master student, Molmed Master, Erasmus MC
Katrin Pachler - Master student, Molmed Master, Erasmus MC
