What we do
About our project
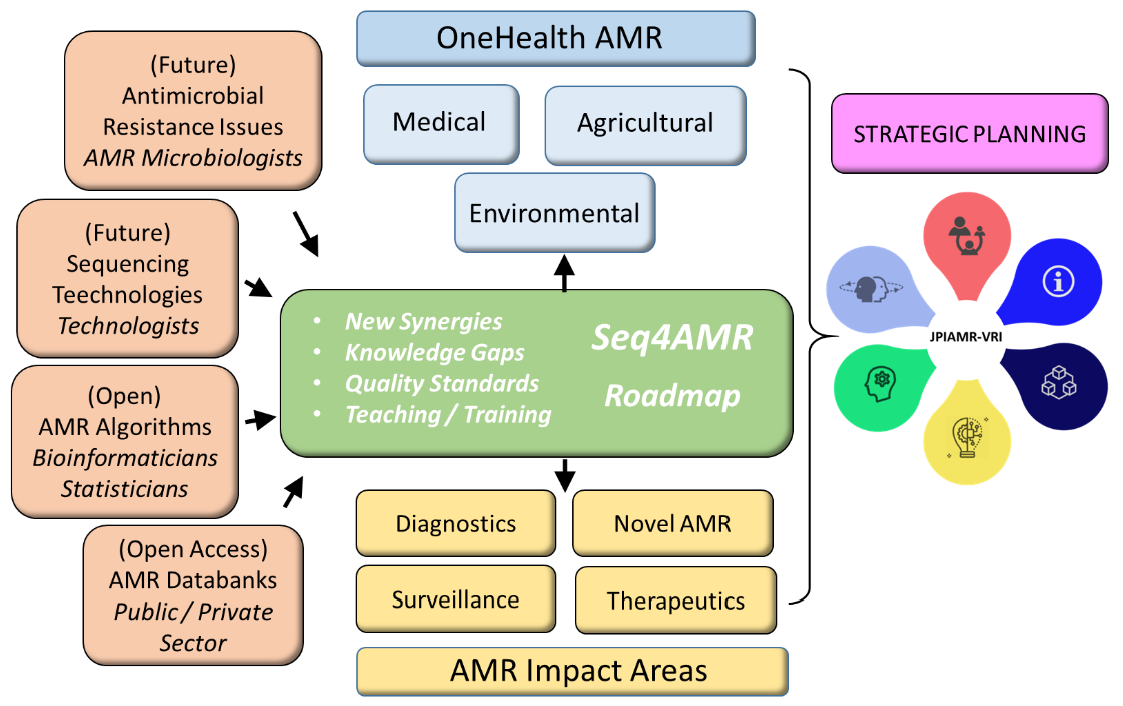
Objectives:
- Promote active collaboration between interdisciplinary OneHealth AMR NGS stakeholders
- Identify knowledge gaps and provide solutions to current/future AMR NGS issues
- Formulate recommendations on quality and quality materials
- Educate AMR NGS stakeholders via interdisciplinary-directed AMR NGS teaching/training materials
Activities:
- 3 Networking Meetings
- Publication of a Seq4AMR Strategic Roadmap
- Open access publications
- Dedicated interdisciplinary Seq4AMR webinar(s) and course(s)
- Dedicated Seq4AMR workshop at an international meeting
Expected Results:
- Establish new OneHealth AMR synergies between international and interdisciplinary experts for knowledge exchange, joint publications grant writing etc.
- Identify current knowledge gaps and how to best fill these gaps
- Formulate quality recommendations and access to materials
- Develop new interdisciplinary AMR teaching/training/materials
- A Seq4AMR Strategic Roadmap
- To contribute and strengthen the activities of the Joint Programming Initiative on Antimicrobial Resistance (JPIAMR)
Our research focus
How can we best identify and promote collaboration between stakeholders who (potentially) utilize next generation sequencing (NGS) to determine the status of antimicrobial resistance (AMR) in bacteria.
By establishing an international and interdisciplinary OneHealth network of public and private experts to identify potential knowledge gaps and solutions for NGS stakeholders (potentially) involved in measuring antimicrobial sensitivity/resistance. Also, by promoting collaboration with other working groups concerned with cross-cutting priorities for NGS and AMR stakeholders.
Funds & Grants
The Seq4AMR network is co-funded by ZonMW (the Netherlands) and the Swedish Research Council (Sweden) as part of the JPIAMR 2020 Networking program.
Collaborations
Internal collaboration:
- Andrew Stubbs, Clinical Bioinformatics Unit, Dept. Pathology
External collaborations:
- Erik Kristiansson, Co-Coordinator, Chalmers University of Technology, Sweden
- Liping Ma, East China Normal University, China
- Willy Valdivia, Orion Integrated Biosciences Inc., USA
- Alex van Belkum, bioMérieux, France
- Sebastian Bruchmann, University of Cambridge, UK
- Andrew McArthur, McMaster University, Canada
- Stefan Emler, SmartGene GmbH, Switzerland
- Eric Claas, Leiden University Medical Centre, The Netherlands
- Andreas Posch, Ares Genetics GmbH, Austria
- Richard Stabler, London School of Hygiene & Tropical Medicine, UK
- Aitana Lebrand, SIB Swiss Institute of Bioinformatics, Switzerland
Our team
- John P. Hays (Co-coordinator) , Dr., Associate Professor
- Astrid Heikema, Dr., Postdoctoral Scientist
- Andrew Stubbs, Dr., Associate Professor
- S. Hiltemann, Dr., Postdoctoral Scientist
