What we do
About our project
Key challenge: global changes in global trends acting as drivers of infectious disease emergence and spread in the One health domain
The digital revolution coupled with data science, and the use of breakthrough multi-analyte technologies offers great opportunities for infectious disease detection. Emerging diseases and antimicrobial resistance (AMR) are increasing due to global changes that have a fundamental impact on the dynamics of infectious diseases, challenging the existing global health infrastructure and organization. Rapid identification is essential to reduce the impact and costs of outbreaks. As a major part of EID and AMR come from animals, a true shift in ability to detect would come from deep understanding the factors that drive their emergence, focusing on the complex interplay of environmental and human factors that drive disease dynamics, and using those insights to develop early warning systems (Figure 1).

Figure. 1: A. VEO’s pro-active, forward looking approach versus the current, reactive approach in EID preparedness and response research (A) and in terms of focusing on drivers of disease emergence and spread instead of taking actions once disease mergence is reported to the healthcare system (B).
VEO will work from the vision that the detection and prediction of human and animal health threats arising from current and predicted global changes can be revolutionized by shifting from a mostly reactive (single) pathogen targeted surveillance (figure 1), into a system that supports:
• prediction, detection and tracking of when and where risk of outbreaks is increasing,
• follow-up of signals for filling data gaps using sophisticated catch-all methodologies and citizen science for verification and tracking
• translating combined data into intuitive and interactive interpretation and visualisation tools for users.
Our research focus
Below, the seven project objectives are described, as summarized in Figure 2. In short, VEO should enable early detection and assessment of changes in drivers that may affect pathogen and AMR circulation. In parallel, VEO will develop tools to integrate high-density data from the newest tools in outbreak research (metagenomics, third generation sequencing, peptide arrays) in risk assessments. We postulate that the integration of these novel types of biodata with contextual (big) data via a range of data mining tools based on advanced analytics will allow detection of changes that increase the risk of outbreaks, with an augmented resolution in time and space, thereby leading to earlier detection and possibly even prediction of outbreaks. The application, added value, and user-friendliness of VEO analytics and tools will be tested in selected scenario’s reflecting the key pathways of emergence for Europe. Outputs of these studies will be used to develop intuitive interpretation software, modelled after the Influenza Pandemic Risk Assessment tool of WHO but bringing in the advanced analytic outputs and visualisations. VEO will use guidance from a panel of stakeholders representing key partner organisations and projects, including the recently established joint group of WHO and the International Telecom Union on artificial intelligence for public health.

Figure. 2: Overview of the envisaged VEO system, enabling handling, integration and analysis of currently used biodata data (light grey box)) and various novel ‘non-traditional’ contextual data sets (dark blue box, top left) for forecasting, nowcasting and tracking of changes in key global drivers of EID emergence and generating actionable information for early warning, risk assessment and monitoring of EID threats. Numbers 1-7 refer to the VEO project objectives.
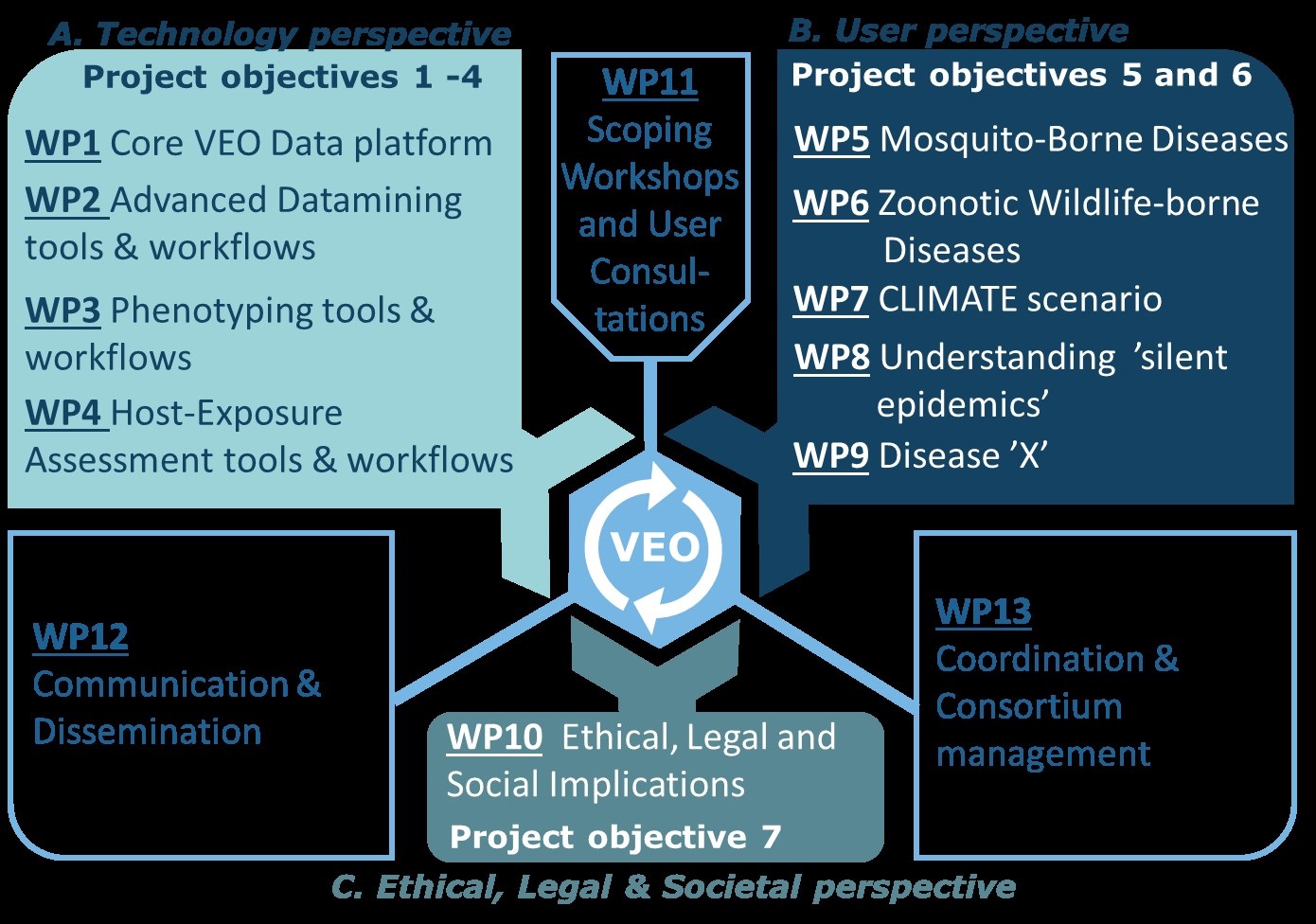
The VEO project is divided into 13 work packages (WP’s, see Figure 3).
• WPs 1 to 4 cover the advanced analytics and technological perspective of objectives 1 to 4 respectively, which are aimed at design and development of core components of VEO.
• WPs 5 to 9 cover the user perspectives of the five use-case scenarios, covering objective 5, and providing key inputs to WPs. 1- 4.
• WPs 10 and 11 cover the societal perspective, with WP10 studying and mapping of the PEARL barriers and WP11 covering the Scoping workshops and User Consultations.
• WP12 includes communication and dissemination activities.
• WP13 completes our WP list with the Consortium management activities.
Funds & Grants
Collaborations
Internal Collaboration:
Viroscience
External Collaborations:
The VEO consortium is highly complementary in terms of European geographic coverage, expertise and disciplinary backgrounds and roles in EID preparedness and response. The VEO consortium consists of 20 partners from 12 European countries, of which 11 member states and one associated country. The broad geographic coverage underscores the true European dimension and relevance of the VEO system and research. With partners from the west, north, south and east of Europe, VEO has good coverage of the different ecosystems, which is particularly relevant for the Use case scenarios research performed in VEO. Moreover, with 12 different countries, VEO has a representative set of health systems and national regulatory landscapes covered.
Participant No |
Participant organization name |
|
1 |
Erasmus University Medical Centre ( Erasmus MC ) |
| 2 | Technical University of Denmark ( DTU ) |
| 3 | Friedrich-Loeffler-Institute ( FLI ) |
| 4 | European Molecular Biology Laboratory ( EMBL ) |
| 5 | Animal and Plant Health Agency ( APHA ) |
| 6 | Eötvös Loránd University ( ELTE ) |
| 7 | Rijksinstituut voor Volksgezondheid en Milieu (RIVM ) |
| 8 | L'Institute Francais de Recherché pour l'Exploitation de la Mer ( IFREMER ) |
| 9 | Aristotle University Thessaloniki ( AUTH ) |
| 10 | Academic Medical Centre ( AMC ) |
| 11 | Institute Pasteur ( IP ) |
| 12 | Consejo Superior de Investigaciones Cientificas ( CSIC ) |
| 13 | École Polytechnique Fédérale de Lausanne ( EPFL ) |
| 14 | Centre for Big data Statistics ( CBS ) |
| 15 | University of Edinburgh ( UEDIN ) |
| 16 | University of Bologna ( UNIBO ) |
| 17 | University of Padova (UNIPD ) |
| 18 | Uppsala University ( UU ) |
| 19 | University of Helsinki ( UH ) |
| 20 | University of Copenhagen ( UCPH ) |
Our team
Project leader:
Professor M.P.G. Koopmans, DVM PhDHead department of Viroscience
Erasmus MC
Wytemaweg 80 - room Ee 1726
3015 CN Rotterdam
Tel: 00 31 10 704 4066
Email: m.koopmans@erasmusmc.nl
Project manager:
Maarten HoekDvM, EPIET, MPH
Program manager
Erasmus MC – Viroscience
Tel: 010 704 40 66 - 41567
Email: m.r.hoek@erasmusmc.nl

