About Prof. Dr. J. (Jurgen) Marteijn, PhD
Introduction

Jurgen Marteijn is professor Transcription stress and DNA damage response at the Molecular Genetics department of the Erasmus MC and PI at the Oncode Institute. The Marteijn lab studies the severe effects of DNA damage on transcription including its physiological consequences like aging. Cells rely on RNA polymerase II (Pol II)-mediated transcription to accurately transcribe their genomes, ensuring proper gene expression and correct cellular function. However, the DNA template that is transcribed by Pol II is continuously challenged by DNA damage, which stall Pol II and disrupt the production of essential RNA molecules. If not properly resolved, these Transcription-blocking DNA lesions (TBLs) result in transcription stress and cellular dysfunction, ultimately leading to neurodegeneration, and premature aging. Specialized transcription-coupled repair (TCR) pathways have evolved that remove TBLs to preserve transcriptional integrity, an essential process underscored by the severe premature aging and developmental and neurological defects observed in patients with inherited TCR deficiencies.
Our lab studies the molecular mechanisms that govern cellular responses to transcription stress and TCR, to uncover how transcription-blocking DNA lesions drive aging and disease. By combining advanced live-cell imaging with state-of-the-art multi-omics approaches, we investigate how cells sense and remove DNA lesions that obstruct transcription, how cells cope with transcription stress and how failures in these pathways contribute to disorders such as Cockayne Syndrome and neurodegeneration.
Over the years, using CRISPR/Cas9 genomic screens and proteomics, we identified several core factors of the transcription-coupled nucleotide excision repair (TC-NER) pathway, including UVSSA, STK19, ELOF1, and HLTF, and demonstrated their molecular mechanisms, shaping the current understanding of TC-NER. Our work also revealed that damage-stalled Pol II is the primary driver of Cockayne Syndrome pathology, providing a unifying explanation for the disease’s severe clinical features. Beyond TC-NER, our lab recently discovered a new transcription-coupled repair pathway dedicated to removing DNA-protein crosslinks (DPCs). We elucidated its mechanism, showed that it acts independently of TC-NER, and linked transcription-blocking DPCs directly to neurodegeneration. Moreover, we discovered of transcriptional responses upon DNA damage involving the spliceosome and R-loops, damage-induced histone exchange, and degradation of promoter-paused Pol II. To enable these discoveries, we developed multiple single-cell tools for sensitively quantifying TCR activity and transcription stress responses in living cells.
Field(s) of expertise
Transcription Stress and Genome Stability
Unperturbed transcription of eukaryotic genes by RNA polymerase II (Pol2) is crucial for proper cell function. However, the DNA template of Pol2 is continuously challenged by damaging agents that can impede or block transcription. These transcription-blocking DNA lesions (TBLs) can cause Genoem stability, cellular dysfunction, senescence and apoptosis, eventually resulting in DNA damage-induced aging. Cells counteract these deleterious effects by transcription-coupled repair (TCR), which specifically removes TBLs thereby safeguarding transcription. My lab uses a combination of state-of-the-art live-cell imaging, advanced proteomic and genomic approaches to identify new proteins involved in this pathway and to obtain a better understanding of the molecular mechanism how cells cope with TBLs.
Research Introduction
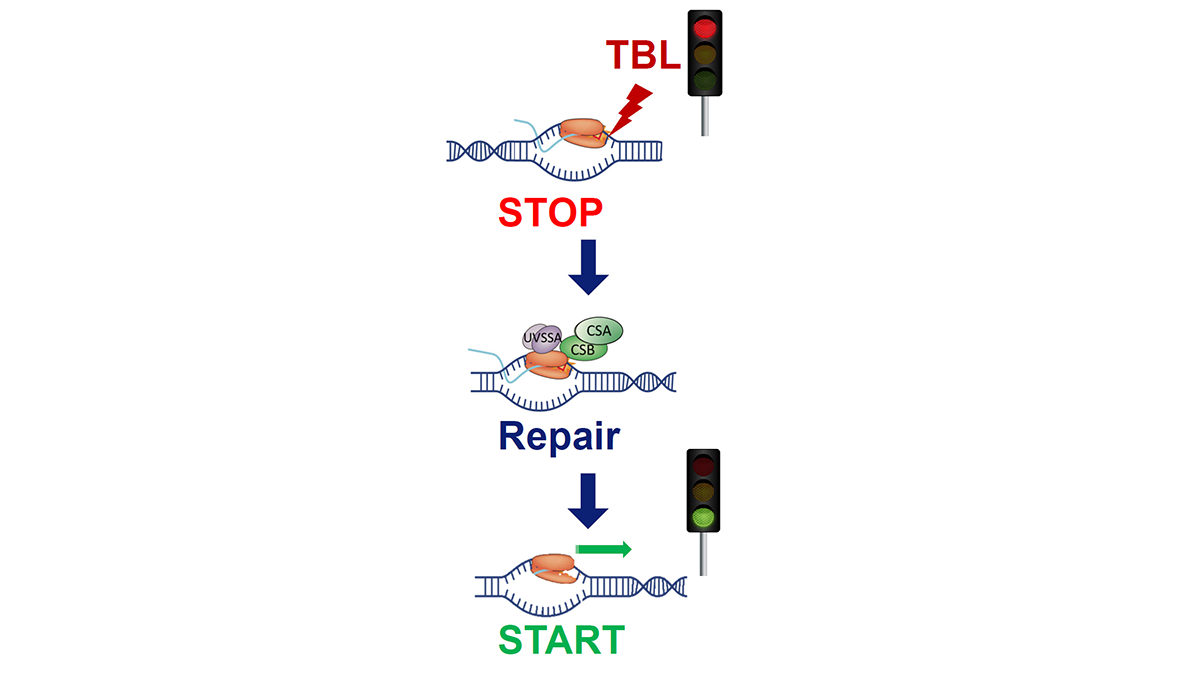
The eukaryotic genome is transcribed by RNA polymerase 2 (Pol 2). Pol 2 mediated transcription is a tightly controlled process to ensure correct temporal and spatial regulation of gene expression, which is crucial for proper cell function. However, the DNA template that is transcribed by Pol 2 is compromised on a daily basis by numerous types of DNA-damaging factors. DNA damage can block or strongly impede Pol 2 mediated transcription. If these transcription-blocking DNA lesions (TBLs) are not properly resolved, this will severely disrupt cellular homeostasis due to the disturbed synthesis of new RNA molecules. In addition, TBLs may result in genome instability, severe cellular dysfunction or premature cell death, which finally may result in DNA damage-induced accelerated aging. Cells counteract these harmful effects by specifically removing TBLs by transcription-coupled repair (TCR), thereby safeguarding transcription. The severe developmental, neurological and premature aging features observed in patients with inherited TCR defects underscore the biological relevance of TCR and show the involvement of TBLs in the aging process.
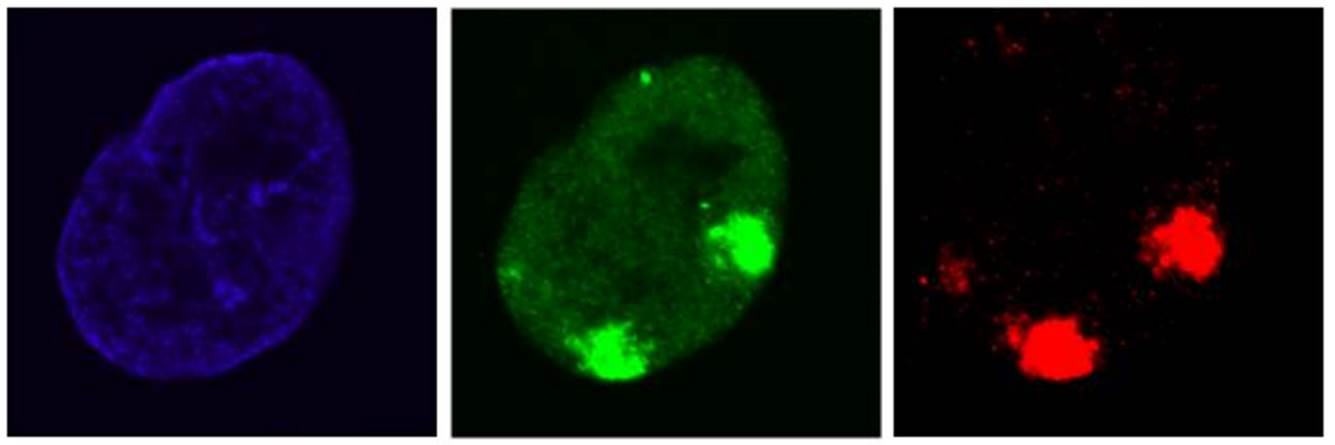
The main research focus of my lab is to investigate the precise regulation and impact of TCR. Over the years, my lab has identified several factors that play important roles in this repair pathway. Although these studies resulted in new insights in the molecular mechanism of the TCR reaction itself, thus far surprisingly little is known about the regulation of transcription or what exactly happens with Pol 2 following the induction of transcription-blocking DNA damage. For example, the exact mechanisms how transcription is inhibited upon DNA damage, or how it is restarted following repair of the TBLs remains largely enigmatic. We use a combination of innovative live-cell imaging approaches with advanced proteomics and genomics approaches to identify new factors involved in the regulation of this pathway and to obtain a better understanding of the molecular mechanisms how cells cope with transcription blocking DNA damage.
DNA Damage and Repair
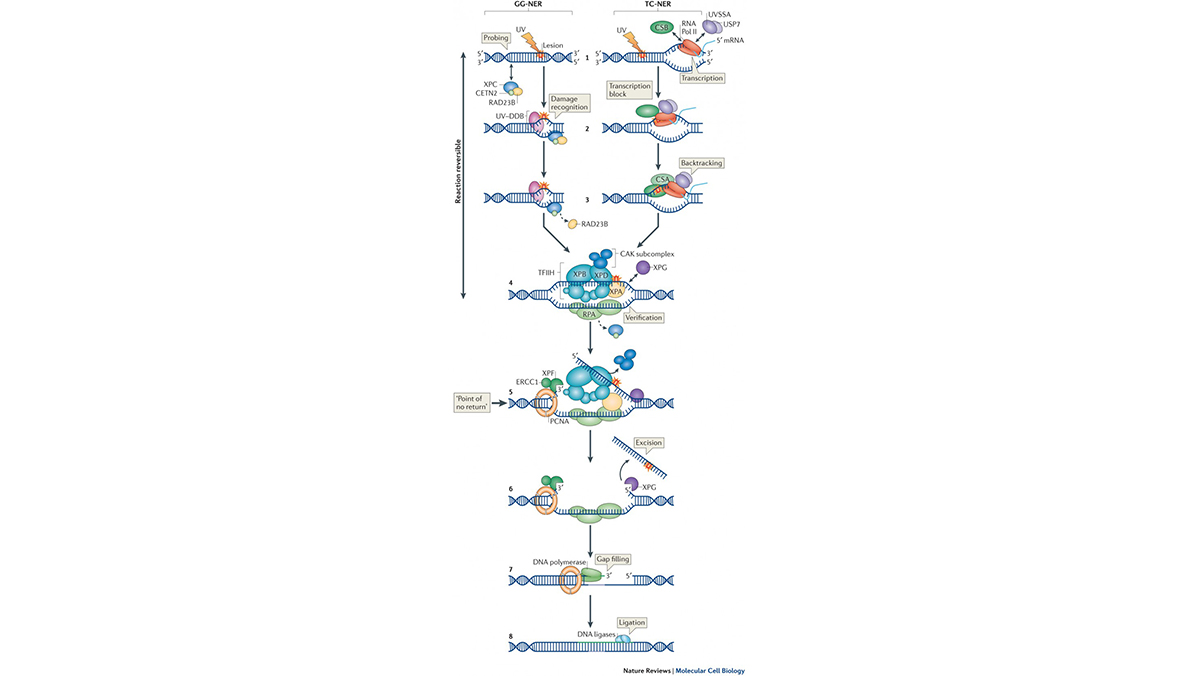
The mammalian genome is protected against genotoxic insults by a network of DNA-damage response (DDR) mechanisms including different DNA repair and damage signaling pathways, to remove lesions and to activate cell cycle checkpoints. Nucleotide Excision Repair (NER) is an important DNA repair mechanism able to remove a broad range of different types of helix-distorting DNA lesions. NER protects organisms against DNA damage-induced carcinogenesis and premature aging. Its significance is illustrated by the severe clinical consequences associated with inherited defects in NER. Genetic defects in NER give rise to various photo-sensitive syndromes, which include the cancer-prone disease Xeroderma Pigmentosum (XP), the premature ageing Cockayne’s Syndrome (CS), and the UV-sensitive syndrome (UVSS).
Transcription blocking DNA lesions (TBLs) cause cellular dysfunction, senescence and apoptosis, finally resulting in DNA damage induced aging. Cells counteract these deleterious effects by transcription-coupled repair (TCR), which removes RNA polymerase 2 (RNAP2) stalling DNA damage. The severe symptoms associated with inherited TCR defects underscore its importance to health but are strikingly diverse, ranging from mild photosensitivity to severe developmental, neurological and premature aging features. Even though TCR was discovered almost 3 decades ago, surprisingly little is known about the molecular consequence and fate of RNAP2 when stalled at different types of TBLs, how transcription is inhibited and subsequently restarted upon repair. Also why TBLs affect tissues differently and how this relates to aging remains enigmatic.
We apply a multidisciplinary approach to improve our understanding of the molecular mechanism of NER and the impact of NER on biological systems, working from the molecular genetic, biochemical and cell biological level to the level of intact eukaryotic organisms and patients.
Selected Publications

- van Sluis, M., Gonzalo-Hansen, C., Li, Q., Lans, H., Wang, D. and Marteijn, J.A.
Mechanisms of transcription-coupled repair and DNA damage surveillance in health and disease.
Nat Rev Mol Cell Biol (2025). 10.1038/s41580-025-00915-3 - van Sluis, M., Yu, Q., van der Woude, M., Gonzalo-Hansen, C., Dealy, S.C., Janssens, R.C., Somsen, H.B., Ramadhin, A.R., Dekkers, D.H.W., Wienecke, H.L., et al. and Marteijn, J.A.
Transcription-coupled DNA-protein crosslink repair by CSB and CRL4(CSA)-mediated degradation.
Nat Cell Biol 26, 770–783 (2024). 10.1038/s41556-024-01394-y - Ramadhin, A.R., Lee, S.H., Zhou, D., Salmazo, A., Gonzalo-Hansen, C., van Sluis, M., Blom, C.M.A., Janssens, R.C., Raams, A., Dekkers, D., et al. and Marteijn, J.A.
STK19 drives transcription-coupled repair by stimulating repair complex stability, RNA Pol II ubiquitylation, and TFIIH recruitment.
Mol Cell 84, 4740–4757.e4712 (2024). 10.1016/j.molcel.2024.10.030
- Gonzalo-Hansen, C., Steurer, B., Janssens, R.C., Zhou, D., van Sluis, M., Lans, H. and Marteijn, J.A.
Differential processing of RNA polymerase II at DNA damage correlates with transcription-coupled repair syndrome severity.
Nucleic Acids Res 52, 9596–9612 (2024). 10.1093/nar/gkae618
- van Toorn, M., Turkyilmaz, Y., Han, S., Zhou, D., Kim, H.S., Salas-Armenteros, I., Kim, M., Akita, M., Wienholz, F., Raams, A., et al. and Marteijn, J.A.
Active DNA damage eviction by HLTF stimulates nucleotide excision repair.
Mol Cell 82, 1343–1358.e1348 (2022). 10.1016/j.molcel.2022.02.020
- Steurer, B., Janssens, R.C., Geijer, M.E., Aprile-Garcia, F., Geverts, B., Theil, A.F., Hummel, B., van Royen, M.E., Evers, B., Bernards, R., et al. and Marteijn, J.A.
DNA damage-induced transcription stress triggers the genome-wide degradation of promoter-bound Pol II.
Nat Commun 13, 3624 (2022). 10.1038/s41467-022-31329-w - Geijer, M.E., Zhou, D., Selvam, K., Steurer, B., Mukherjee, C., Evers, B., Cugusi, S., van Toorn, M., van der Woude, M., Janssens, R.C., et al. and Marteijn, J.A.
Elongation factor ELOF1 drives transcription-coupled repair and prevents genome instability.
Nat Cell Biol 23, 608–619 (2021). 10.1038/s41556-021-00692-z - Wienholz, F., Zhou, D., Turkyilmaz, Y., Schwertman, P., Tresini, M., Pines, A., van Toorn, M., Bezstarosti, K., Demmers, J.A.A. and Marteijn, J.A.
FACT subunit Spt16 controls UVSSA recruitment to lesion-stalled RNA Pol II and stimulates TC-NER.
Nucleic Acids Res 47, 4011–4025 (2019). 10.1093/nar/gkz055
- Steurer, B., Turkyilmaz, Y., van Toorn, M., van Leeuwen, W., Escudero-Ferruz, P. and Marteijn, J.A.
Fluorescently-labelled CPD and 6-4PP photolyases: new tools for live-cell DNA damage quantification and laser-assisted repair.
Nucleic Acids Res 47, 3536–3549 (2019). 10.1093/nar/gkz035 - Lans, H., Hoeijmakers, J.H.J., Vermeulen, W. and Marteijn, J.A.
The DNA damage response to transcription stress.
Nat Rev Mol Cell Biol 20, 766–784 (2019). 10.1038/s41580-019-0169-4 - Steurer, B., Janssens, R.C., Geverts, B., Geijer, M.E., Wienholz, F., Theil, A.F., Chang, J., Dealy, S., Pothof, J., van Cappellen, W.A., Houtsmuller, A.B. and Marteijn, J.A.
Live-cell analysis of endogenous GFP-RPB1 uncovers rapid turnover of initiating and promoter-paused RNA Polymerase II.
Proc Natl Acad Sci U S A 115, E4368–E4376 (2018). 10.1073/pnas.1717920115
- Wienholz, F., Vermeulen, W. and Marteijn, J.A.
Amplification of unscheduled DNA synthesis signal enables fluorescence-based single-cell quantification of transcription-coupled nucleotide excision repair.
Nucleic Acids Res 45, e68 (2017). 10.1093/nar/gkw1360 - van Cuijk, L., van Belle, G.J., Turkyilmaz, Y., Poulsen, S.L., Janssens, R.C., Theil, A.F., Sabatella, M., Lans, H., Mailand, N., Houtsmuller, A.B., Vermeulen, W. and Marteijn, J.A.
SUMO and ubiquitin-dependent XPC exchange drives nucleotide excision repair.
Nat Commun 6, 7499 (2015). 10.1038/ncomms8499 - Tresini, M., Warmerdam, D.O., Kolovos, P., Snijder, L., Vrouwe, M.G., Demmers, J.A., van I.W.F., Grosveld, F.G., Medema, R.H., Hoeijmakers, J.H., Mullenders, L.H., Vermeulen, W. and Marteijn, J.A.
The core spliceosome as target and effector of non-canonical ATM signalling.
Nature 523, 53–58 (2015). 10.1038/nature14512 - Dinant, C., Ampatziadis-Michailidis, G., Lans, H., Tresini, M., Lagarou, A., Grosbart, M., Theil, A.F., van Cappellen, W.A., Kimura, H., Bartek, J., Fousteri, M., Houtsmuller, A.B., Vermeulen, W. and Marteijn, J.A.
Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage.
Mol Cell 51, 469–479 (2013). 10.1016/j.molcel.2013.08.007 - Schwertman, P., Lagarou, A., Dekkers, D.H., Raams, A., van der Hoek, A.C., Laffeber, C., Hoeijmakers, J.H., Demmers, J.A., Fousteri, M., Vermeulen, W. and Marteijn, J.A.
UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair.
Nat Genet 44, 598–602 (2012). 10.1038/ng.2230



