About Dr. N. (Nitika) Taneja, PhD
Introduction
Dr. Nitika Taneja, PhD is an Associate Professor and Principal Investigator at the Department of Molecular Genetics, Erasmus University Medical Center (Erasmus MC) and also affiliated with Technical University, Delft (TU, Delft) since 2018. She is an Oncode Investigator and consequently her group became part of Oncode Institute since 2024. The central theme of her research activities has been focused on understanding complex molecular mechanisms of chromatin remodeling upon DNA replication and replication stress in maintaining genome stability.
"Since replication stress is one of the main causes of inducing DNA damage upon chemotherapy treatment, dr. Taneja is exploring the potential role of histone modifications and chromatin remodeling pathways involved in re-organizing chromatin locally & spatially to develop tolerance towards replication stress and acquiring chemoresistance in cancer cells."

She obtained her PhD from the University of Zurich (2013), where she studied transgenerational inheritance of histone variants in Drosophila. As a postdoctoral fellow at the National Cancer Institute, NIH (2017), she investigated heterochromatin inheritance during DNA replication in fission yeast.
In her own lab, she employs mammalian systems, including human and mouse cell lines, tumor organoids, and patient-derived material (ovarian and prostate cancer samples such as ascites, circulating tumor cells, and biopsy-derived cells), to study chromatin organization at replication forks, particularly in the context of chemotherapy-induced replication stress. Her work addresses a major knowledge gap in understanding how three-dimensional chromatin architecture mediates replication fork stability under stress. By uncovering these connections and identifying the key molecular factors involved, her research opens new avenues in the fields of cancer and disease. To this end, her group develops state-of-the-art single-molecule and 3D genomics tools to overcome the limitations of conventional methods, enabling investigation of transient chromatin changes both locally at replication forks and at the genome-wide level to capture spatial reorganization of chromatin under replication stress.
She has been recognized with several prestigious grants and awards, including the Young Investigator Award from the Daniel den Hoed Stichting Foundation, Erasmus+ Fellowship, Convergence Open Mind Call, NWO-Vidi Talent Award, Aspasia Premium Grant, NWO Recognition Award and Incentive Grant for Women in STEM, and a European Research Council (ERC) Starting Grant. She was also selected through a nationwide competitive process as a Junior Oncode Investigator, supported by the Oncode Institute.

Field(s) of expertise
3-D Chromatin Remodeling & Replication
Our lab investigates local and spatial re-organization of chromatinized replication forks in response to replication stress. We use a combination of super-resolution
imaging, genomics and proteomics-based approaches to understand the complex mechanisms of chromatin regulation in maintaining replication fork stability in normal as well as cancer cells.
We uses mammalian system (human/mouse cell lines, tumor organoids and patient tumor samples including PDX) to study chromatin organization upon replication stress.
1. Nucleosome remodeling at forks
We have described a hitherto unknown mechanism, by which the SWI/SNF chromatin remodeler, SMARCAD1 stabilizes active replication forks, independently of its role in DNA repair, that is essential to maintaining resistance towards replication poisons. We further show how fork protection-challenged BRCA1-deficient naïve- or chemoresistant tumors require SMARCAD1-mediated active fork stabilization to maintain unperturbed fork progression essential for tumor (cells and organoids) viability. Lo et al. 2021, Science Advances.
2. Dynamics of histone modifications at forks
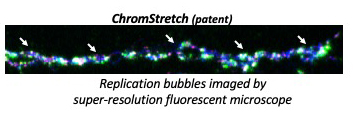
We are further investigating the role of epigenetic signatures and chromatin modifiers in initiating the chromatin signaling cascade in response to replication stress and their checkpoint-dependent regulation in distinguishing the two states of fork dynamics, using our innovative super-resolution microscopy-based and proteomics technologies. ChromStretch technology (Patent 2022).

3. R-loop homeostasis in vicinity of forks
We are studying the enigmatic role of RNA:DNA hybrids in facilitating epigenetic landscape and the potential role of replication fork preservation pathways, and the assisting Chro-Mates (chromatin modifiers and remodelers) in their removal further avoiding replication‐transcription conflicts. Uruci et al. 2021, IJMS.
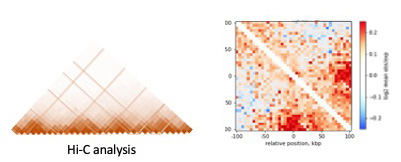
4. 3-D chromatin re-organization upon replication stress
Spatial re-organization of replication forks: We are investigating how local and spatial re-organization of replicating chromatin regulates replication stress response in cells. We are continuously developing novel tools to observe the 3-D chromatin organization using specialized single molecule to next-generation deep sequencing-based technologies.
Publications
Publications

- Bakker C†, Paulson J†, Taneja N. Fencing the genome: ubiquitin signaling restricts heterochromatin spread. (Signal Transduction and Targeted Therapy, Nature/articles/s41392-025-02349-x, 2025)
- Galloy M, Blodeau A, Vion E, Kim D, Bakker CA, Gaggioli C, Thomas M, Lavoie EG, Marois I, Monterroso ADD, Masson JY, Taneja N, Miller K, Maréchal A, Fradet-Turcotte A. Ubiquitination of the histone variant mH2A1.2 prevents toxic RAD18 accumulation at a subset of genomic loci upon replication stress. (Molecular Cell 2025, PMID: 40812313)
- Cherdyntseva V, Paulson J, Adakli S, Gagné, Aouami, Ubieto-Capella, González-Acosta C, Bakker C, Poirier G, Taneja N, Lopes M. Nucleoplasmic Lamin A/C controls replication fork restart upon stress by modulating local H3K9me3 and ADP-ribosylation levels. (Nature Communications 2025, Accepted. BioRxiv https://doi.org/10.1101/2025.01.18.633737)
- Lingeman J, Heuvel Dvd, Berg Jvd, Singh JK, van Hooijdonk R, Wiegant WW, Boer DEC, Lo SY, Panagopoulos A, Gonzalez-Prieto R, Franken M, Kolsters N, de Jong OG, Vertegaal ACO, Altmeyer M, Taneja N, van Oudernaarden A, Luijsterburg M, Kasri NN, Latour B, van Attikum H. The NSL complex promotes neural development by preventing R-loop induced replication stress. (BioRxiv, DOI: 10.1101/2025.08.22.671281)
- Lo CSY, Taneja N*, Ray Chaudhuri A*. Enhancing quantitative imaging to study DNA damage response: A guide to automated liquid handling and imaging. (DNA repair 2024, DOI: 10.1016/j.dnarep.2024.103769)
- Uruci S, Hoitsma N, Solér-Oliva E, Bayona-Feliu A, Gaggioli V, García-Rubio, Lo CSY, Bakker C, Marinello, Manolika EM, Capranico G, Luijsterburg M, Luger K, Aguilera A, Taneja N. SMARCAD1 Regulates R-Loops at Active Replication Forks Linked to Cancer Mutation Hotspots. (BioRxiv https://doi.org/.09.13.612941)
- Davó-Martínez C, Helfricht A, Ribeiro-Silva C, Raams A, Tresini M, Uruci S, van Cappellen WA, Taneja N, Demmers JAA, Pines A, Theil AF, Vermeulen W, Lans W. Different SWI/SNF complexes coordinately promote R-loop- and RAD52-dependent transcription-coupled homologous recombination. (Nucleic Acid Research 2023, DOI: 10.1093/nar/gkad609)
- Gaggioli V, Lo CSY, Reverón-Gómez N, Jasencakova Z, Domenech H, Nguyen H, Sidoli S, Tvardovskiy A, Uruci S, Slotman JA, Chai Y, Goncalves JG, Manolika EM, Jensen ON, Wheeler D, Sridharan S, Chakraborty S, Demmers J, Kanaar R, Groth A, Taneja N. Dynamic de novo heterochromatin assembly and disassembly at replication forks ensure fork stability. (Nature Cell Biology 2023, DOI: 10.1038/s41556-023-01167-z)
- This article was selected for News & Views in Nature Cell Biology by Susan M. Gasser. Transient chromatin compaction in fork restart. (Nature Cell Biology 2023, DOI: 10.1038/s41556-023-01181-1)
PATENTS
DUTCH NATIONAL PATENT #NL-EP-PCT/P132000EP00. Methods, compositions and devices for spreading of chromatin fibers.
Lead Inventor: Nitika Taneja, Co-inventors: Vincent Gaggioli, Roland Kanaar
My Group

The Chromatin Remodeling and Replication group.




